

Sterols and hopanoids are sometimes used as biomarkers to infer the origin of certain groups of organisms. Traditionally, hopanoid-derived products in ancient rocks have been considered to indicate the presence of bacteria, whereas sterol derivatives have been considered to be exclusive to eukaryotes. However, a closer look at the topic reveals a rather complex distribution of either compound in both bacteria and eukaryotes. (1). The known biosynthetic pathways for sterols and hopanoids are similar but diverge at a critical step where two different enzymes are used: squalene-hopene cyclase (SHC) and oxidosqualene cyclase (OSC), the latter requiring oxygen. These two enzymes belong to the same gene family, whose complex evolutionary history is difficult to reconcile with the known species phylogeny.
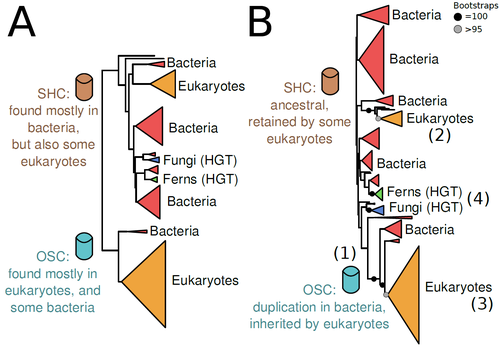
In this study (2), Dr. Warren R. Francis revisits the evolution of this gene family using an extended dataset with a broader taxonomic representation. In contrast to the traditional representation of the tree rooted between SHC and OSC paralogs (i.e., based on function), the author proposes that rooting the tree within bacterial SHCs and assuming a secondary origin of OSC is more parsimonious. This postulates SHC to be the ancestral function –retained in many extant bacteria and some eukaryotes– and OSC to have emerged later within bacteria –currently being mostly present in eukaryotes–. The reconstructed evolutionary history is arguably complex and can only be reconciled with the species' phylogeny by invoking many secondary losses. These losses are considered likely because many extant species acquire sterols and hopanoids by diet and lack one or both enzymes. Some cases of recent horizontal gene transfer are also proposed.
In contrast to the dichotomy between bacterial SHCs and eukaryote OSCs, the new proposed scenario suggests that the eukaryote ancestor likely inherited both enzymes from bacteria and thus could be able to synthesize both sterols and hopanoids. Under this hypothesis, not only bacteria but also eukaryotes could be responsible for the hopane found in old rocks. This agrees with eukaryote fossils dating back to more than 1 billion years ago (3). Also, the observed increase of sterane levels in rocks ~600-700 million years old cannot be associated with the origin of eukaryotes, which is a much older event, but could rather reflect changes in atmospheric oxygen levels because oxygen is required for the synthesis of sterols by OSC.
References
1. Santana-Molina C, Rivas-Marin E, Rojas AM, Devos DP (2020) Origin and Evolution of Polycyclic Triterpene Synthesis. Molecular Biology and Evolution, 37, 1925–1941. https://doi.org/10.1093/molbev/msaa054
2. Francis WR (2022) The Eukaryotic Last Common Ancestor Was Bifunctional for Hopanoid and Sterol Production. Preprints, 2020040186, ver. 5 peer-reviewed and recommended by Peer Community in Evolutionary Biology. https://doi.org/10.20944/preprints202004.0186.v5
3. Butterfield NJ (2000) Bangiomorpha pubescens n. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology, 26, 386–404. https://doi.org/10.1666/0094-8373(2000)026<0386:BPNGNS>2.0.CO;2
DOI or URL of the preprint: https://doi.org/10.20944/preprints202004.0186.v4
Version of the preprint: 4
Dear Dr. Francis,
Thank you for submitting your new revised manuscript. I agree with the reviewers that this new version is much improved after most of the previous concerns were addressed. Reviewers have provided some additional comments that still require clarification, but these are rather specific and therefore easy to address.
One of the reviewers suggest some methods for tree rooting, which you might want to explore. This might provide information on the specific bacterial branch where the root might be and thus could strengthen your study, but I do not think this is a requirement. I would urge you, however, to double check your writing is always as careful as it should, especially when explaining your evolutionary hypothesis, which relies on a tree root that makes sense but has basically no support from data. For example, in the abstract “suggests” might be better than “indicates” and “is easier explained” would be better than “is best explained”. Caption of figure 2 and page 5: “SHC probably was the ancestral enzyme” rather than “SHC was the original enzyme”.
Minor comments: In page 3, “The latter three of these make sense with respect to the tree.” You refer to the known species phylogeny and not to Fig 1, right? Please, also check the text throughout for small typos, I found a few but was difficult to note without line numbers.
I like this manuscript and I am close to recommend it. However, this new version is not that different from the previous one, about which I had made a number of comments. The new figures are welcome and the revised text is clearer at several places. Yet there are still a couple ambiguous phrasings and topics for discussion that should at least be mentioned. Here is a list of past and new comments:
Major Comments
Minor Comments
Typos (I provide context since there are no line numbers)
This is the second revision I make to the manuscript entitled “The eukaryotic last common ancestor was bifunctional for hopanoid and sterol production”, by Dr. Francis. First of all, I would like to apologise to Dr. Francis and Dr. Irisarri for the delay in my response. I was curious about the corrected version and wanted to participate in this second round, but the timing ended up being not so good for me.
Despite the many comments, suggestions, and corrections made during the first round, I think the author has done an excellent job addressing all the concerns, and has provided a thorough response to all of them. I do believe the manuscript has improved from the previous version. It might be because I have already read it before, but I find it easier to read and to understand. I’m recommending the acceptance of the manuscript.
However, it is worth mentioning that the major issue remains: the strength of the hypothesis depends on how reliable the rooting of the tree is. I am not very familar with these methods, but I wonder if some method designed to root phylogenetic trees without an outgroup (such as STRIDE, Emms and Kelly, 2017, MBE: https://doi.org/10.1093/molbev/msx259) could help with this. Likewise, not many weeks ago it was published the “rootstrap” method, aiming to assess the support for a given root in the tree (Naser-Khdour et al., 2021, Syst. Biol.: https://doi.org/10.1093/sysbio/syab067). Maybe they do not identify the “correct” placement of the root, but they might provide extra support to the hypothesis presented. This is more a suggestion than a request.
In any case, I acknowledge there is little that can be done on this regard and the author already addresses this issue correctly in the main text, but I also think this should be included in the recommendation that would be published in parallel to the manuscript. Future readers might find it useful.
https://doi.org/10.24072/pci.evolbiol.100371.rev32Dear,
I consider that the author has contributed to improve the manuscript with the comments. However, I would like to emphasize in one of them. I mentioned that the methods about the trimming process for the matrix should be incorporated into the manuscript so I recommend to the author to add his reply to the method section.
https://doi.org/10.24072/pci.evolbiol.100371.rev33
DOI or URL of the preprint: https://doi.org/10.20944/preprints202004.0186.v2
Version of the preprint: 2
Dear Dr. Francis,
Thank you very much for submitting your revised manuscript. It might seem like the responses to the third reviewer's comments are missing from the uploaded point-by-point letter, could you please check this before we consider your preprint?
Thank you very much for your time. Sincerely,
Iker Irisarri
DOI or URL of the preprint: https://doi.org/10.20944/preprints202004.0186.v1
Version of the preprint: 1
Dear Dr. Francis,
Thank you for your submission and apologies for the delay in our response; it took some time to collect all the reviews.
You will see from the Reviewers’ comments and my own that we definitely think there is merit in your work, but before this preprint can be recommended some revisions would be needed.
As noted by one of the reviewers, one key publication in MBE covers several important aspects that are directly relevant to this preprint. It puts the evolution of sterol/hopanoid pathways in a broader context and tackles several points that are of relevance for your work, and therefore should be appropriately discussed.
A recurrent concern seems to be tree rooting, which is neither trivial nor arbitrary. One of the Reviewers suggests an interesting reference and the MBE paper by Santana-Molina et al. discusses also this topic. Could the presence of SHC/OSC homologs in Archaea help in this task? This possibility is not mentioned by the author, but I assume archaean homologs are unlikely based on what is said in Santana-Molina et al.? Regarding the hypothesis in Fig 1B, I also agree that the bacterial root probably makes most sense. However, the hypothesized evolutionary history of these enzymes rests on this particular rooting for which evidence seems to be scarce besides the phylogenetic distribution of paralogs that seem quite complex anyway. As pointed out by the Reviewers, this assumption should be made explicit and the real uncertainty of the hypothesized scenario better reflected by revising the tone of the relevant sections. The uncertainty could also be better reflected in other sections of the manuscript as well, such as when discussing biomarkers in fossils.
From my viewpoint, the manuscript contains too many expressions that e.g., personalize molecules, assign “choices” to organisms, or claim “bizarre” loss events. Such expressions fit very well into what has been called the “night science” language (Yanai & Lercher 2019; https://doi.org/10.1186/s13059-019-1800-6) but should be avoided in scientific publications. Note also Reviewers’ comments on specific sections where the writing could be improved for clarity and correctness. Finally, there are a few demonstrative pronouns without an object following them (“this”), which sometimes make it difficult to follow the argumentation (e.g. point 2.3).
Regarding the general structure of the paper, I think the current “narrative” structure helps transmitting the Author’s view, but the data behind the claims are not always clear. For example, when laying out the evolutionary history in points 2.2.1 to 2.2.6, it could help to refer to specific parts of the tree that back up each claim, thereby better connecting the phylogenetic hypothesis and the biological interpretation.
I appreciate the transparency of centrally depositing alignments and trees in Bitbucket. It might be good to add a fully annotated tree as a supplement, given that the current taxon names in the tree short and not very informative, and only a cartoon of the tree is presented in the manuscript. Fig. 1 needs support values (at least for the key nodes) and perhaps a rewrite of the caption because the current one “Recreated tree from [Takishita et al., 2012]” might sound like the tree correspond to the old tree by Takishita et al. Note also the comment that asks about how the new added sequenced improved our view on the story. Could the data of Santana-Molina et al. MBE paper change that? Please, take into account the considerations on phylogenetic methodology raised by two of the Reviewers. I wondered whether using HMM or psiBLAST could help identify more distant homologs that might be relevant. For BLAST searches, significance thresholds should be provided.
Lastly, one of the Reviewers suggests adding a schematic figure setting up the stage for the evolutionary questions. This might clarify one doubt I had: Fig 1 of Santana-Molina et al. depicts SQMO as a step in the sterol but not in the hopanoid synthesis, whereas in the preprint I understand that this oxygen-dependent step of SQMO is common to both hopanoid and sterol production: “Following the oxygen-dependent step, one of two enzymes then forms the multi-ring structure, either squalene-hopene cyclase (SHC) for hopanoids or oxidosqualene cyclase (OSC) for sterols.” Which one is right or am I missing something here?
The study of ancient organisms represents a major challenge in evolutionary biology, as the evidence is scarce and many hypotheses rely on sporadical observations. The development of phylogenetic methods has become an important addition to this field, allowing the use of extant species in comparative analyses under an historical perspective: if most of the lineages of a clade share a given trait, it is very likely that trait has been inherited from a common ancestor. Here, in the manuscript entitled “The eukaryotic last common ancestor was bifunctional for hopanoid and sterol production”, Dr. Francis uses such methods to explore the origin of the production of hopanoids and sterolds, two important membrane elements.
I really appreciate the simplicity of the methods, enough to answer the question proposed. Thus far, the analysis of the results and the presentation of the hypothesis are clear and well written, which makes this manuscript easy to read. I would like to congratulate the author for this effort. However, I do have one major concerns that is related to the general tone of the manuscript. Here, the author proposes a new hypothesys for the origin of SHC and OSC enzymes. After reading the manuscript a couple of times, I always have the feeling that the author presents this new hypothesys (LECA already had both enzymes) as a fact based on the evidence, but that is not true. I like this hypothesis and I think that is very likely, based on the results presented, but all the body of evidence relies in one simple assumption: that the tree must be rooted in one particular branch. Even if that rooting makes sense based in our current knowledge, the lack of an outgroup is a major burden because rooting the tree in a different branch might end up in a different story and we cannot disregard that possibility. This limitation is of course addressed in the main text, but I think the manuscript should never leave that “hypothetical ground”, if I may say so. To make this point clear: “an extended dataset suggests the presence of SHC and OSC in the LECA”, instead of “LECA already presented SHC and OSC”. Overall, I suggest the acceptance of this manuscript.
I also have some minor suggestions that the author might want to consider: - In general, I find confusing the use of so many acronyms along the text. SHC, STC, OCT, SQMO, HGT… I understand the use of these acronyms, it’s just that sometimes it requires an extra effort to understand what the author is talking about. - I wonder if hopanoids and steroids appear in the same rocky substrates and, if not, if there is evidence of one of them in more ancient rocks. In the conclussion the need of a thorough review of the fossil record is mentioned, and I agree that this would be a really valuable addition to this problem. - Page 7, “some bacteria appear have”: appear to have. - Page 8, “the genomes of cnidarians (corals and jellyfish), ctenophores”: and ctenophores.
https://doi.org/10.24072/pci.evolbiol.100371.rev11The manuscript of W.R. Francis deals with an interesting problem: the origin and evolution of hopanoid/sterol biosynthetic pathways and especially of triterpene cyclase enzymes (SHC/STC and OSC).
I don't have strong feelings against this specific piece of work, except the fact that it was released one month after the publication of a large study covering the same topic (and more) by Devos et al. in Mol Biol Evol (2020) [doi:10.1093/molbev/msaa054]. Therefore, I think that the latter study should be mentioned and its findings discussed in the present manuscript to be really useful to the community.
My other comments are mostly minor and meant to clarify some points. Here is the list below.
- The introduction is just a bit too brief to fully understrand the study for someone not familiar with the pathways at play. Indeed, Fischer and Pearson (2007) do not provide an overview of the two pathways nor of their interplay, whereas Nes (2011) is too technical for the argument here while providing no details about hapanoids. Figure 1 in Devos et al. (2020) is much better in this respect, but still overly complex for what is needed here. Please consider adding a schematic figure (or panel to an existing figure) to efficiently set up the scentific background. This would also allow the author to precisely define what is STC, which is not yet achieved in the current version of the manuscript.
- I am convinced that the author knows what he is discussing. However, I am sometimes puzzled by the vocabulary. For example, this sentence is ambiguous to me: "All else being equal, the presence of OSC in some bacteria and stem eukaryotes is nonetheless best explained by primary inheritance of OSC by a pre-eukaryotic host from a bacterial endosymbiont." Similarly, "This is explained by vertical inheritance from bacteria at the origin of eukaryotes (probably endosymbiosis)" and "primary inheritance in the LECA from a bacterial endosymbiont at the origin of mitochondria" look confusing. In my opinion, it is important to distinguish the multiple possible cases here. Even if a pathway (or enzyme) is ancestral to all extant eukaryotes (i.e., present in LECA) and then vertically spread, it can be there for multiple reasons: 1) ancestral to all three domains (vertical evolution), 2) present in the archaeal host cell (if there was any such thing, again vertical evolution), 3) provided by a bacterial symbiont (e.g., the future mitochondrion, thus endosymbiosis), 4) "invented" along the eukaryotic stem (i.e., ESP, none of the above). Alternatively, the pathway (enzyme) can be introduced into a subset of extant eukaryotic lineages through H/LGT (horizontal/lateral evolution). These options should be clarified upfront. Regarding case (3), it may be useful to mention that, according to Devos et al. (2020), a mitochondrial origin of OSC is unlikely ("The scarcity of OSC in Alpha-proteobacteria indicates that this bacterial contribution is unlikely to be related to the mitochondria").
- Another example is the lengthy discussion about rooting (split into multiple parts in the manuscript). While I of course agree with the fact that most phylogenetic reconstructions do not yield rooted trees, I am less fond of the claim that rooting is necessarily an "arbitrary decision". What is required is external evidence, and indeed distinct functions in case of multigene families can be such evidence. Here, I have the impression that ancestrally duplicated genes (i.e., prior to the evolution of the three domains) appear unlikely to the author ("this implies a parallel origin of the two enzymes relative to the unknown outgroup"). Considering that such genes do exist, I think this position should be better argued. For some ideas about rooting the tree of life, I humbly refer to our own piece: Gouy et al. (2015) in Philos Trans R Soc Lond B Biol Sci [doi: 10.1098/rstb.2014.0329].
- Somewhat in opposition to the previous comment, Devos et al. (2020) argue that "the current data do not provide enough resolution to infer the presence of SHC in a common bacterial ancestor, although the results do indicate that the biosynthesis of hopanoid (defined by the SHC enzyme) precedes the diver- sification of the whole Gracilicutes group and some of the Terrabacteria taxa". This should be kept in mind for the first step of the scenario developed in the present study ("SHC was the original enzyme, distributed across many bacterial lineages"). Related to that, the following sentence is confusing ("nor is it clear which bacterial lineages may have possessed this enzyme due to horizontal gene transfer between prokaryotic lineages"). For me, the question is not well formulated: either SHC is ancestral to all existing bacteria and there is no point discussing extant lineages or it appeared in a lineage that was already distinct from other still-extant lineages and thus not ancestral to all. What is the author's hypothesis? Furthermore, the sentence about OSC emergence by duplication of SHC (or a SHC/OSC ancestor I would rather say if I reasoned as a pure cladist) draws a parallel with the distribution of SHC that is unjutified to me ("it is also currently unclear who this might be", emphasis mine) because OSC is much less extensively distributed among bacteria and thus the question of the lineage of origin appears more legitimate than for SHC.
- Co-emergence and/or co-evolution of OSC with SQMO ("It is very likely that this was coincident with the origin of SQMO") would make sense but is not strictly required since all these enzymes seem to exhibit some level of metabolic plasticity (see Devos et al., 2020). In the present work, SQMO distribution and evolution is not discussed at all. So it is difficult to take a stance about this issue. Related to that, understanding the "choice made in each [eukaryotic] lineage to keep either STC/SHC or OSC" would require to answer the following two questions: Can STC work after SQMO? Can OSC work without SQMO? All these points should be better discussed based on the current literature. In this respect, the explanatory ideas about the similarly patchy distribution of elongation factors (EF1a/EFL) might come handy (e.g., Keeling and Inagaki, 2004, PNAS; doi: 10.1073/pnas.0404505101).
- Regarding Methods and Figures, I think that they can be improved. Figure 1B should differentiate at least some bacterial groups if the origins of OSC (and eukaryotic SHC/STC) have to be discussed (as it appear to be the case in the main text, e.g., "Fungi have acquired SHC from an ancestor of Anaeromyxobacter, a delta-proteobacterium"). Moreover, some idea of the statistical support at important nodes is required to understand if there is phylogenetic resolution at all. For example, the dimensions of the alignment should be provided. Related to that, how were identified the MMETSP sequences (by TBLASTN searches against mRNA sequences or by BLASTP against predicted proteins)? How did the author deal with the contaminant sequences plaguing some samples of the original MMETSP dataset?
https://doi.org/10.24072/pci.evolbiol.100371.rev12