

 based on reviews by Rubén González and 2 anonymous reviewers
based on reviews by Rubén González and 2 anonymous reviewers
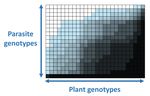
In a landmark paper, Flores et al. (2011) showed that the interactions between bacteria and their viruses could be nicely described using a bipartite infection networks. Two quantitative properties of these networks were of particular interest, namely modularity and nestedness. Modularity emerges when groups of host species (or genotypes) shared groups of viruses. Nestedness provided a view of the degree of specialization of both partners: high nestedness suggests that hosts differ in their susceptibility to infection, with some highly susceptible host genotypes selecting for very specialized viruses while strongly resistant host genotypes select for generalist viruses. Translated to the plant pathology parlance, this extreme case would be equivalent to a gene-for-gene infection model (Flor 1956): new mutations confer hosts with resistance to recently evolved viruses while maintaining resistance to past viruses. Likewise, virus mutations for expanding host range evolve without losing the ability to infect ancestral host genotypes. By contrast, a non-nested network would represent a matching-allele infection model (Frank 2000) in which each interacting organism evolves by losing its capacity to resist/infect its ancestral partners, resembling a Red Queen dynamic. Obviously, the reality is more complex and may lie anywhere between these two extreme situations.
Recently, Valverde et al. (2020) developed a model to explain the emergence of nestedness and modularity in plant-virus infection networks across diverse habitats. They found that local modularity could coexist with global nestedness and that intraspecific competition was the main driver of the evolution of ecosystems in a continuum between nested-modular and nested networks. These predictions were tested with field data showing the association between plant host species and different viruses in different agroecosystems (Valverde et al. 2020). The effect of interspecific competition in the structure of empirical plant host-virus infection networks was also tested by McLeish et al. (2019). Besides data from agroecosystems, evolution experiments have also shown the pervasive emergence of nestedness during the diversification of independently-evolved lineages of potyviruses in Arabidopsis thaliana genotypes that differ in their susceptibility to infection (Hillung et al. 2014; González et al. 2019; Navarro et al. 2020).
In their study, Moury et al. (2021) have expanded all these previous observations to a diverse set of pathosystems that range from viruses, bacteria, oomycetes, fungi, nematodes to insects. While modularity was barely seen in only a few of the systems, nestedness was a common trend (observed in ~94% of all systems). This nestedness, as seen in previous studies and as predicted by theory, emerged as a consequence of the existence of generalist and specialist strains of the parasites that differed in their capacity to infect more or less resistant plant genotypes.
As pointed out by Moury et al. (2021) in their conclusions, the ubiquity of nestedness in plant-parasite infection matrices has strong implications for the evolution and management of infectious diseases.
References
Flor, H. H. (1956). The complementary genic systems in flax and flax rust. In Advances in genetics, 8, 29-54. https://doi.org/10.1016/S0065-2660(08)60498-8
Flores, C. O., Meyer, J. R., Valverde, S., Farr, L., and Weitz, J. S. (2011). Statistical structure of host–phage interactions. Proceedings of the National Academy of Sciences, 108, E288-E297. https://doi.org/10.1073/pnas.1101595108
Frank, S. A. (2000). Specific and non-specific defense against parasitic attack. Journal of Theoretical Biology, 202, 283-304. https://doi.org/10.1006/jtbi.1999.1054
González, R., Butković, A., and Elena, S. F. (2019). Role of host genetic diversity for susceptibility-to-infection in the evolution of virulence of a plant virus. Virus evolution, 5(2), vez024. https://doi.org/10.1093/ve/vez052
Hillung, J., Cuevas, J. M., Valverde, S., and Elena, S. F. (2014). Experimental evolution of an emerging plant virus in host genotypes that differ in their susceptibility to infection. Evolution, 68, 2467-2480. https://doi.org/10.1111/evo.12458
McLeish, M., Sacristán, S., Fraile, A., and García-Arenal, F. (2019). Coinfection organizes epidemiological networks of viruses and hosts and reveals hubs of transmission. Phytopathology, 109, 1003-1010. https://doi.org/10.1094/PHYTO-08-18-0293-R
Moury B, Audergon J-M, Baudracco-Arnas S, Krima SB, Bertrand F, Boissot N, Buisson M, Caffier V, Cantet M, Chanéac S, Constant C, Delmotte F, Dogimont C, Doumayrou J, Fabre F, Fournet S, Grimault V, Jaunet T, Justafré I, Lefebvre V, Losdat D, Marcel TC, Montarry J, Morris CE, Omrani M, Paineau M, Perrot S, Pilet-Nayel M-L and Ruellan Y (2021) The quasi-universality of nestedness in the structure of quantitative plant-parasite interactions. bioRxiv, 2021.03.03.433745, ver. 4 recommended and peer-reviewed by PCI Evolutionary Biology. https://doi.org/10.1101/2021.03.03.433745
Navarro, R., Ambros, S., Martinez, F., Wu, B., Carrasco, J. L., and Elena, S. F. (2020). Defects in plant immunity modulate the rates and patterns of RNA virus evolution. bioRxiv. doi: https://doi.org/10.1101/2020.10.13.337402
Valverde, S., Vidiella, B., Montañez, R., Fraile, A., Sacristán, S., and García-Arenal, F. (2020). Coexistence of nestedness and modularity in host–pathogen infection networks. Nature ecology & evolution, 4, 568-577. https://doi.org/10.1038/s41559-020-1130-9
DOI or URL of the preprint: https://www.biorxiv.org/content/10.1101/2021.03.03.433745v1
 , posted 10 May 2021
, posted 10 May 2021Dear Dr. Moury et al.,
Sorry for the long time it took me to gather three opinions on your manuscript. As you will see, all three reviewers find the paper very interesting and that, eventually, will represent a nice contribution to the field of virus ecology. However, all three also consider that the manuscript needs substantial editions before it can be acceptable, both in terms of structure, clarification of certain points, and adding missing methodological information. I'm sure you'll find their comments very useful.
Yours,
See attached pdf
Download the reviewReview Moury et al.
In this study the authors want to understand the shape of interaction matrices between hosts and their parasites. Two features are of importance, nestedness and modularity. These are measured in 32 matrices of quantitative pathogenicity trait data gathered from 15 plant-parasite pathosystems consisting of either annual or perennial plants along with fungi or oomycetes, bacteria, nematodes, insects and viruses. After assessing the performance of several nestedness and modularity algorithms by simulation (in SI Material), they find that significant modularity in only six of the 32 matrices, with two or three modules detected. Significant nestedness is found in 30 of the 32 matrices. Interestingly, nestedness was linked to a parasite strain effect and a plant accession effect, with no parasite-plant interaction term.
I found the paper very interesting, very well written and easy to follow. I am convinced by the analyses, and as far as I can check, the statistical analyses are robust and well conducted. The interpretation of the results is cautious and takes into account the uncertainties and statistical limitations of the methods. I therefore highly recommend this study for PCI Evol Biol, and have only some minor points to improve the clarity of the paper, or indicating some missing information, and links to the theoretical literature as suggestions.
Introduction:
Three topics could be better explained:
1) It would be nice to link nestedness and modularity to the general models by Boots (Boots et al. 2014 Evolution for example). These models allow very general switch from quantitative to qualitative traits based on underlying trade-offs and simple assumptions on the interaction matrix.
2) The role of epistasis between genes of interaction could be mentionned. I would expect epistasis to generate modularity (but that is a guess). In animals several papers by the group of Dieter Ebert do show some example of epistatic interactions of GxG interactions. In plants, one could imagine the same for multi-locus plant-parasite model (e.g. in a multilocus GFG model with quantitative value, i.e. not 0/1, see for example papers by Sasaki).
3) The link to non-host resistance and parasite effectors to overcome this resistance is also not clear, would we expect some modularity then?
- Line 97: Older citations would be more appropriate than Thrall et al. 2016 (for example papers by Gandon and/or Nuismer who have already made this explicitly). There is a nice paper by Dybdhal et al. (2014 Am Nat) which is also very relevant. Some definitions used are found in Antonovics et al. (2012, Evolution) which present some points of the introduction in a general way for plant and parasite systems.
Results and Methods:
- Line 173: It is unclear whether in all systems, the minimum and maximum of disease severity are comparable (as the scale is then set to be the same for all system between 0 and 9). Does this mean that all systems have reported the value 0 as no infection at all, and value 9 as maximum disease severity?
This clarification is needed to understand how quantitative or qualitative are the disease severity measures in the different systems.
- Line 183, as well line 774-777: it would be nice to add some information on the null models (some names are given but the reader cannot assess what they mean without diving in the SI Material). It would be nice to indicate which null model test is the most stringent and which one is the most relaxed. A general description of the main null models and their assumptions (randomization procedure) could be included in the methods. The description is very good in the SI Material but lacks in the text. The description of some infection experiment could be reduced if these are published already to gain space for describing the null models.
- Line 218: should it be “wine” instead of “wNODF”? otherwise this sentence is confusing.
Discussion:
- Line 441: The paper by Fenton, Antonovics and Brockhurst (Evolution 2012) also generates via a two-step infection process (similar to non-host resistance followed by infectivity/resistance) some interesting predictions. I guess that there modularity and/or nestedness generated by such model can be also predicted and applicable to plant systems?
- I missed a discussion of the difference between annual/perennial and between different types of pathogens (Oomycetes, virus, bacteria, nematodes,...) on why nestedness or modularity should be expected to be found. Is there a link between nestedness/modularity with life history traits such as obligate or facultative parasitism?
- What could be expectations for nestedness/modularity for wild plant systems versus crops? Could these make a nice set of predictions to be tested in other systems as conclusion of the paper?
The manuscript entitled “The quasi-universality of nestedness in the structure of quantitative plant-parasite interactions” studies plant-pathogen interaction quantitative data by analyzing the structure of 32 matrices. The authors evaluated multiple algorithms, using them to analyze the structure and nestedness of the matrices. The findings point to a universal scarce modularity and high nestedness in the plant-parasite interactions. The strengths of the paper consist in the test of multiple algorithms, the use of a diverse set of plant-parasites, and the study of the structure of quantitative data. The authors have nicely described the methodology and datasets used, following a clear logic for their analysis. The examination of the biological significance of modularity and nestedness outcomes contributes to better understand the implications of the results. The analysis done uses a limited set of plant-parasites but, considering the scarce number of available data, the work makes relevant progress to indagate in the shared structures of plant parasite-interactions. This manuscript will be relevant for scientists working in the areas of plant disease, host-microbes interactions, and evolution.
Comments:
1. In the discussion section there is no comment on the suitability of the algorithms used. The wNODF and WINE algorithm are discussed in the results sections. In the introduction it is said that one of the two goals of the work is "to assess the performance of available algorithms" (L152-153), but in the conclusion section there is no statement about the best suited algorithms.
2. L264-265 The main text misses one reference when citing the algorithms: for label prop algorithm (Raghavan et al., 2007).
3. L366-367 It is said that from the 32 matrices only two of them are not nested. The text speculates that the one of the two non-nested matrices could be nested but its little size may be hindering the nested pattern. As the size of the matrices were decided before the analysis, the results should be trusted.
4. L510-511 Datasets were select under the criteria of having at least 6 plant accessions and 6 parasite strains. Is this minimum number arbitrary or was it required for the application of the algorithms?
5. In Figure 2 all matrices are shown and the significant nested ones are numbered in red. Some identification for the matrix that showed modularity for infection or resistance scores would be appreciated.
6. For the Figure 3 it is stated that coefficient of correlation could not be calculated in some cases (L254-257). How many times did this happen? The figure is nice but I could not find the raw data in order to observe the coefficient value for each matrix depending on the threshold.