

Polyploidy, which results in the presence of more than two sets of homologous chromosomes represents a major feature of plant genomes that have undergone successive rounds of duplication followed by more or less rapid diploidization during their evolutionary history. Polyploid complexes containing diploid and derived polyploid taxa are excellent model systems for understanding the short-term consequences of whole genome duplication, and have been particularly well-explored in evolutionary ecology (Ramsey and Ramsey 2014, Rice et al. 2019). Many polyploids (especially when resulting from interspecific hybridization, i.e. allopolyploids) are successful invaders (te Beest et al. 2012) as a result of rapid genome dynamics, functional novelty, and trait evolution. The origin (parental legacy) and modes of formation of polyploids have a critical impact on the subsequent polyploid evolution. Thus, elucidation of the genomic composition of polyploids is fundamental to understanding trait evolution, and such knowledge is still lacking for many invasive species.
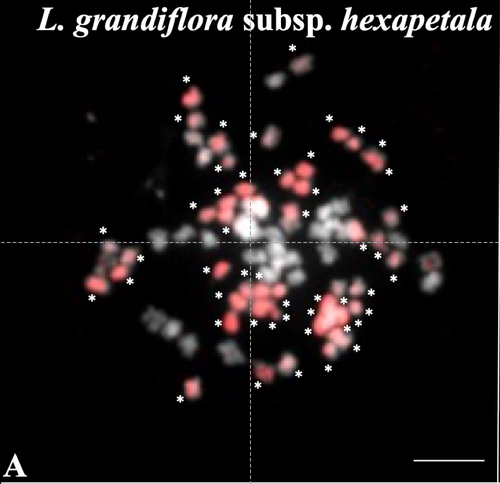
Genus Ludwigia is characterized by a complex taxonomy, with an underexplored evolutionary history. Species from section Jussieae form a polyploid complex with diploids, tetraploids, hexaploids, and decaploids that are notorious invaders in freshwater and riparian ecosystems (Thouvenot et al.2013). Molecular phylogeny of the genus based on nuclear and chloroplast sequences (Liu et al. 2027) suggested some relationships between diploid and polyploid species, without fully resolving the question of the parentage of the polyploids. In their study, Barloy et al. (2023) have used a combination of molecular cytogenetics (Genomic In situ Hybridization), morphology and experimental crosses to elucidate the genomic compositions of the polyploid species, and show that the examined polyploids are of hybrid origin (allopolyploids). The tetraploid L. stolonifera derives from the diploids L. peploides subsp. montevidensis (AA genome) and L. helminthorhiza (BB genome). The tetraploid L. ascendens also share the BB genome combined with an undetermined different genome. The hexaploid L. grandiflora subsp. grandiflora has inherited the diploid AA genome combined with additional unidentified genomes. The decaploid L. grandiflora subsp. hexapetala has inherited the tetraploid L. stolonifera and the hexaploid L. grandiflora subsp. hexapetala genomes. As the authors point out, further work is needed, including additional related diploid (e.g. other subspecies of L. peploides) or tetraploid (L. hookeri and L. peduncularis) taxa that remain to be investigated, to address the nature of the undetermined parental genomes mentioned above.
The presented work (Barloy et al. 2023) provides significant knowledge of this poorly investigated group with regard to genomic information and polyploid origin, and opens perspectives for future studies. The authors also detect additional diagnostic morphological traits of interest for in-situ discrimination of the taxa when monitoring invasive populations.
References
Barloy D., Portillo-Lemus L., Krueger-Hadfield S.A., Huteau V., Coriton O. (2024). Genomic relationships among diploid and polyploid species of the genus Ludwigia L. section Jussiaea using a combination of molecular cytogenetic, morphological, and crossing investigations. BioRxiv, ver. 4 peer-reviewed and recommended by Peer Community in Evolutionary Biology https://doi.org/10.1101/2023.01.02.522458
te Beest M., Le Roux J.J., Richardson D.M., Brysting A.K., Suda J., Kubešová M., Pyšek P. (2012). The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany, Volume 109, Issue 1 Pages 19–45, https://doi.org/10.1093/aob/mcr277
Ramsey J. and Ramsey T. S. (2014). Ecological studies of polyploidy in the 100 years following its discovery Phil. Trans. R. Soc. B369 1–20 https://doi.org/10.1098/rstb.2013.0352
Rice, A., Šmarda, P., Novosolov, M. et al. (2019). The global biogeography of polyploid plants. Nat Ecol Evol 3, 265–273. https://doi.org/10.1038/s41559-018-0787-9
Thouvenot L, Haury J, Thiebaut G. (2013). A success story: Water primroses, aquatic plant pests. Aquat. Conserv. Mar. Freshw. Ecosyst. 23:790–803 https://doi.org/10.1002/aqc.2387
DOI or URL of the preprint: https://doi.org/10.1101/2023.01.02.522458
Version of the preprint: 2
First, we would like to address many thanks to recommender and reviewers for their constructive and high-quality comments which greatly brush up the article and once again to Malika Ainouche for accepting to be recommender and organizing an efficient and constructive peer-review.
Concerning corrections requested by the recommender, we rephrased all unclear sentences as requested, changed the words as suggested and verified that Ludwigia was in italic all over the document. The new paragraph about GISH method was placed in corresponding method part.
This revised manuscript has been substantially improved and has addressed all the questions and comments raised by the reviewers. The study provides significant contribution to our undrestanding of the species relationships and the origins of the polyploid species of the section Jussiaea (genus Ludwigia). The text needs however, some English checking and grammar editing as indicated in the comments on the attached edited version. Accordingly, I would recommend this paper after minor revision (without second round of peer review).
Download recommender's annotationsDOI or URL of the preprint: https://doi.org/10.1101/2023.01.02.522458
Version of the preprint: 1
Dear Dr Barloy
Your manuscript « Genomic relationships among diploid and polyploid species of the genus Ludwigia L.,section Jussiaea using a combination of cytogenetic, morphological, and crossing investigations” has been examined by two reviewers, and both agree that this study reports new interesting findings regarding the origins of polyploid Ludwigia species from section Jussiaea, that include notorious intercontinental invaders. The conducted approach and method are adequate, and the study is well designed. However, the reviewers also raised some limitations and provided suggestions that we ask you to take into account before considering the manuscript for publication in PCI. These concerns are mainly (but not exclusively) related with (i) the morphological study, and its relationship with the central question that is answered using cytogenetic investigations, (ii) the structure and clarity of the text.
In addition to the comments of the two reviewers, the manuscript should be also improved as following:
Introduction:
When presenting species of section Jussiaea, previous findings regarding phylogenetic relationships between diploid and polyploid species (e.g. Liu et al. 2017), previous hypotheses regarding auto or allopolyploid origin are missing; this information is provided later in the discussion, but should be presented in the introduction to more clearly highlight the unresolved questions and the need for additional investigations. Similarly, the way the species are circumscribed and the need for useful diagnostic morphological traits to distinguish the species should be mentioned to better justify the morphological analyses that have been undertaken. As it stand the link between the morphological study and the goals of the paper, is not clear
Material and Methods:
Plant material: The sample size (number of individuals) indicated per species is not consistent with the number of individuals examined across the different analyses (morphology, flow cytometry, GISH): please indicate to which analyses the numbers indicated in the plant material refer.
Morphological traits: Please justify the choice of the analyzed traits and the goals of this analysis.
Genome size estimation by flow cytometry:
L. 175: Please provide the genome size of the species used as internal standard (Trifolium and Zea)
Also in these polyploid species it would be better to use “1C” as “gametic” genome size (n), rather than “haploid”, which may be confused with “monoploid” (x). In the results (table 1) 2C values (instead of C-values) are provided
Results
Morphology: lack of statistical analysis (any data regarding variability within species?)
Discussion: Please start with an introductive sentence recalling questions and approaches
Ludwigia species in section Jussiaeae: I would have presented the diagnostic morphological traits (li. 338-345) in either the introduction or the method section to justify the choice of the studied traits.
The sentence l. 347 stating that the morphometric approach is “comprehensive” should be moderated as it is here based on two traits only without statistical analysis across populations over the range of the species (see also comments of Reviewer 1)
Origin of polyploids : rely with their native and introduced range that are not indicated
Line 392: “phylogenetic study”: rather use “genomic relationships and origins of polyploids”
Line 393: we propose the “first phylogenetic history”: this is not appropriate: you could indicate instead “first hypotheses regarding diploid-polyploid relationships”…
When referring to previously published phylogenetic work using nuclear and chloroplast DNA markers: can you discuss the maternal inheritance of cp DNA information compared to biparental ITS-Waxy information and the present findings using GISH ( any comments on the maternal progenitor?).
The section on “combination of different data to identify phylogenetic relationships” is not convincing and not warranted as it stands … The interest of combining different lines of data and the critical contribution of molecular cytogenetics (which cannot be presented as an alternative to other approaches) could be briefly mentioned in the concluding paragraph.
This manuscript reports several genetic analyses of Ludwigia sp. having different ploidy levels with the aim to establish their relationships and to understand the origin of hexa and decaploids. The authors infer a genealogy of species based on their genomic composition revealed by GISH. Genome size measurement, morphological observation and experimental crosses are also used to confirm the characteristics and limits of the taxa. The work which has been done is adequate with the main objectives. Comprehensive cytogenetic studies are rather rare although essential to decipher the origin and evolution of polyploids. Here they offer a different view of evolutionary relationships among taxa than the molecular phylogenetic studies. Such data and results will be very precious for future genomic studies. In my opinion there no major issues in the Ms, however, the structure of the text and the text itself could be improved to facilitate its understanding.
Firstly, the authors should explain, or remind the reader, that with GISH methods, the inference is based on the blocking DNA rather than the probe.
Secondly, the role of morphological observations is unclear as the authors report many features but without any morphometric analysis (measurement or coding of discrete variables and multivariate analyses). Without these morphometric analyses, it is difficult to assess the effects and even the purpose of the morphological observations. Perhaps the authors could use it as a simple verification step to prove that the identification is correct, this would allow them to reduce the size of their text.
Thirdly, the inference by GISH of the origin of the two 4X is clear but it is less clear for the 6X and 10X, the authors need to give more explanation on the relevance of intensity variation (page 13).
Finally, we are left wanting more from the experiment cross, perhaps a figure summarizing which are the crosses that always led to a dead-end could be informative to make hypotheses on what may have happened. Currently, a simple, fast reading may lead to the inference that all the crosses failed, but that’s not true.
Minor concerns :
page 2, l. 44 : so what is diploid if all plants have experienced at least one polyploidy event ?
page 5 l. 117: the word phylogeny should be kept for inference based on phylogenetic methods (e.g. cladistic). Genomic relationships or genealogy is better here.
page 8 l. 191 : how many genomic DNA ?
page 9 l. 222 : does it mean that the experimental crosses were done after GISH and based on GISH inferences ?
page 10 l. 227 : did you used a hood on flower to prevent non-controlled pollination ?
Figure 1 : improve quality of the pictures, it is pixelized.
Table 1 : estimator of variance are needed (Sd or 95% CI)
GISH results : The explanation of the diploid GISH is confusing. I do not understand why and how Lpm and Lh could be “genetically close” but could “correspond to different genomes”. Perhaps I miss it but the overall strategy leading to table 2 should be explained. For example why not blocking the 4X La genome by the 2X Lpm genome ?
Page 14, l. 329 : “no plants survived at 90 days after seedling” : is it due to their genomic composition or to another (external) factor ? do you compare the survival of seedlings among inter and intra specific crosses ?
P 15 l. 349 and after : it’s true that morphometrics done on plants grown in common garden are very precious however morphometrics require measurement or at least standardized observation of discrete variables and analyses (e.g discriminant analysis). For example, the differences between Lh and Lpm need to be confirmed by the data.
Page 20 l. 475-478 : Although I agree on the fact that cytogenetics is crucial, it does not replace molecular phylogenetics or phylogenomics. Both are complementary and both are expensive if the salary costs are considered.
https://doi.org/10.24072/pci.evolbiol.100645.rev11The paper is generally well designed, the structure and origin of the polyploid complex is approached using combination of multiple methodical approaches. There are, however, several issues in the manuscript that should be addressed while preparing its revised version.
(1) The authors should adopt a single taxonomic concept that should be referred to throughout the paper. The current version, where the same taxon is in one paragraph referred to as L. grandiflora subsp. grandiflora and in the other one as L. grandiflora (or in one as L. grandiflora subsp. hexapetala and in the other one as L. hexapetala) is very confusing.
(2) When counting species and subspecies one should not call all of them species (the term “taxa” instead of “species” would be more appropriate). E.g., “one hexaploid species (2n=6x=48) (Ludwigia grandiflora subsp. grandiflora); and one decaploid species (2n=10x=80) (Ludwigia grandiflora subsp. hexapetala)” [lines 85-87] – these are not two separate species, but two subspecies of the same species, i.e., two taxa. The same problem is here: ”It is not easy to distinguish between the hexaploid and decaploid species morphologically and both have previously been treated as a single species (Ludwigia uruguayensis (Cambess.) H. Hara; Zardini et al., 1991)” [lines 87-89] – in the concept adopted in the previous sentence, these are two subspecies of the same species.
“Taxon name” should be used instead of “Species name” should be used also in the Table 1.
(3) Line 117: “phylogenic origin” – what is the exact meaning of this term? Would just “origin” be sufficient?
(4) The description of methods of Chromosome counting, Genome size estimation by flow cytometry, and Genomic in situ hybridization is too long, it can be shortened, providing the reference to some other papers. On the other hand, the description of morphological measurements is not sufficient. The number and origin (locality) of measured plants for each taxon and morphological character should be provided (to document the representativeness of the measurements).
(5) line 142: “Morphological observations for each species were randomly made” – this is unclear (meaning of “randomly”??).
(6) Locality details (at least geographical coordinates of the locality of origin) and number of analyzed plants should be provided for each chromosome number count and genome size measurement for each taxon.
(7) Lines 225 (“produced flowers in continuous on a shoot”), 228 (“for each of other species”) – unclear meaning (language).
(8) Lines 404, 421: “hexaploid” instead of “hexaploidy”.
https://doi.org/10.24072/pci.evolbiol.100645.rev12