

There are many organisms that are asexual or have unusual modes of reproduction. One such quasi-sexual reproductive mode is androgenesis, in which the offspring, after fertilization, inherits only the entire paternal nuclear genome. The maternal genome is ditched along the way. One group of organisms which shows this mode of reproduction are clams in the genus Corbicula, some of which are androecious, while others are dioecious and sexual. The study by Vastrade et al. (2022) describes population genetic patterns in these clams, using both nuclear and mitochondrial sequence markers.
In contrast to what might be expected for an asexual lineage, there is evidence for significant genetic mixing between populations. In addition, there is high heterozygosity and evidence for polyploidy in some lineages. Overall, the picture is complicated! However, what is clear is that there is far more genetic mixing than expected. One possible mechanism by which this could occur is 'nuclear capture' where there is a mixing of maternal and paternal lineages after fertilization. This can sometimes occur as a result of hybridization between 'species', leading to further mixing of divergent lineages. Thus the group is clearly far from an ancient asexual lineage - recombination and mixing occur with some regularity.
The study also analyzed recent invasive populations in Europe and America. These had reduced genetic diversity, but also showed complex patterns of allele sharing suggesting a complex origin of the invasive lineages.
In the future, it will be exciting to apply whole genome sequencing approaches to systems such as this. There are challenges in interpreting a handful of sequenced markers especially in a system with polyploidy and considerable complexity, and whole-genome sequencing could clarify some of the outstanding questions,
Overall, this paper highlights the complex genetic patterns that can result through unusual reproductive modes, which provides a challenge for the field of population genetics and for the recognition of species boundaries.
References
Vastrade M, Etoundi E, Bournonville T, Colinet M, Debortoli N, Hedtke SM, Nicolas E, Pigneur L-M, Virgo J, Flot J-F, Marescaux J, Doninck KV (2022) Substantial genetic mixing among sexual and androgenetic lineages within the clam genus Corbicula. bioRxiv, 590836, ver. 4 peer-reviewed and recommended by Peer Community in Evolutionary Biology. https://doi.org/10.1101/590836
DOI or URL of the preprint: https://doi.org/10.1101/590836
Version of the preprint: 2
This preprint is suitable for a recommendation but the second reviewer makes a number of suggestions which I would like to give the authors a chance to respond to. In particular further attempt to make the discussion more general and more concise would be an improvement.
The authors have gone to great lengths to address the reviewer comments, including performing additional cloning, reanalysis of all the data, and re-writing most of the manuscript. My main concerns with the previous version were the exclusion of singletons and the interpretations of distinct origins in the Discussion. Both of these have been addresed: singletons are now included, and the Discussion provides a very thouightful and cautious interpretation of results, with a call for better sampling and the use of genome-wide data in the future.
Although this study is an exploration of one unusual genus, it has broader relevance in that it reveals the complex ancestries that can arise through atypical reproductive modes, and the challenges in species delimitation that can result. I therefore think the manuscript will be relevant to many biologists, and that it is suitable for publication.
https://doi.org/10.24072/pci.evolbiol.100198.rev21
The extensive revision has successfully addressed the majority of the points raised during the previous round of reviews. Especially the introduction is now aimed at a much more general readership, and most of the technical issues regarding data and analyses have been clarified. A few points might be need further attention in an additional minor revision.
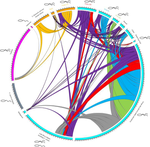
First, the distinction between mitochondrial and nuclear markers could be taken even further. In particular the Circos plot (Fig. 4) apparently mixes nuclear and mitochondrial markers and therefore two very distinct genetic mechanisms: Sharing of mitochondrial alleles between lineages can be explained by androgenetic males using eggs from distinct lineages, whereas sharing of nuclear alleles is evidence for hybridization/nuclear capture. Perhaps consider presenting two circus plots, one for the mitochondrial and the other for the nuclear alleles, so that the two could directly be compared. Of course, hybridization/nuclear capture could also lead to sharing of mitochondrial alleles, so that the comparison of the figures does not directly present a comparison of the two mechanisms. This would need to be explained in more detail.
Second, the discussion is still rather system-specific. It is easy to become lost in the many lineages and comparisons discussed in great detail (and also with some repetition). I think the attractiveness of the preprint to a general readership could be improved by placing stronger emphasis on the major points and by adding a general conclusion that circles back to the main questions raised in the introduction.
A few additional minor comments:
- Initially, the description of androgenesis (L. 59-60) sounds as if a reductional meiosis takes place during spermatogenesis.
- The statement that males or hermaphordites “hijack” eggs of other individuals (L.64) is at odds with the statement that all androgens are hermaphrodites and that they can use their own eggs. Also, given the modified meiosis during oogenesis, which leads to eggs without any chromosomes, I do not completely follow the notion of “parasitism” (at least not during androgenetic “self-fertilization”): These eggs could not be used for any other form of reproduction, so the “parasite” appears to only confer benefit rather than harm to the “host”. Is there an added value to the manuscript of raising the issue of egg parasitism?
- On L. 85-87 it is unclear how the orientation of the meiotic axis can lead to the formation of two polar bodies in a single meiotic division. Either explain better or discard (the argument doesn’t appear to be crucial here).
- There appears to be a contradiction between the first two sentences of the paragraph starting on L 117 (the first sentence says that sexuals occur in Africa, Asia, Australia, and Middle East, the second that they are geographically very restricted).
- L. 154: unclear if “self-fertilize” refers to androgenesis within hermaphrodites or to sexual self-fertilization.
- L. 433: “Fig 4” (instead of “Fig. 3”).
- Perhaps it is worth mentioning that nulear capture/hybridization events are probably much more easily detected if they occur between distant lineages than within lineage. (The latter could be severely under-detected or go un-noticed althogether).
https://doi.org/10.24072/pci.evolbiol.100198.rev23
DOI or URL of the preprint: https://doi.org/10.1101/590836
Version of the preprint: 1
The reviews generally accept the broad interest of the paper and the advances, and I agree with this. However, there are some important comments on the paper that need to be addressed. The key comments include: 1) the framing of the paper and making it more appealing to a general audience - it is currently written very much about the Corbicula clams, but it is unclear how it relates to evolutionary processes on other organisms. This needs work both in the introduction and perhaps also in the conclusions - how does the unique reproductive mode affect the genetic variation in these organisms compared to other systems? 2) Clear statement of the hypotheses being tested. This is particularly important - the final paragraph of the introduction focusses on methods used (i.e. Haplowebs) but does not really outline what the paper is trying to test. Could there be a list of specific hypotheses laid out in this paragraph? 3) Clarify nomenclature such as 'egg parasitism' - I agree with the reviewer that this seems a misleading term and request that a better term is devised. 4) Better clarify what is novel in this paper relative to previous work on the same system. There are a number of references to previous studies but it should be clarified which data comes from previous papers (and if published data are not included why was this the case) and which questions can be addressed here that were not addressed before. More specifically, what exactly is novel here apart from the use of a web based method of analysis. I am also concerned about some of the methodological questions raised by reviewers. How many clones were sequenced per individual and why were triploid allele patterns not detected - is this just because too few clones were sequenced. Also clarify the 'remove singletons' option in the Haploweb program. Why were singletons removed? There are a large number of additional minor suggestions that should be addressed in the reviews. Subject to addressing these points I think the manuscript is appropriate for a recommendation. **Additional requirements of the managing board**: As indicated in the 'How does it work?’ section and in the code of conduct, please make sure that: -Data are available to readers, either in the text or through an open data repository such as Zenodo (free), Dryad (to pay) or some other institutional repository. Data must be reusable, thus metadata or accompanying text must carefully describe the data. -Details on quantitative analyses (e.g., data treatment and statistical scripts in R, bioinformatic pipeline scripts, etc.) and details concerning simulations (scripts, codes) are available to readers in the text, as appendices, or through an open data repository, such as Zenodo, Dryad or some other institutional repository. The scripts or codes must be carefully described so that they can be reused. -Details on experimental procedures are available to readers in the text or as appendices. -Authors have no financial conflict of interest relating to the article. The article must contain a "Conflict of interest disclosure" paragraph before the reference section containing this sentence: "The authors of this preprint declare that they have no financial conflict of interest with the content of this article." If appropriate, this disclosure may be completed by a sentence indicating that some of the authors are PCI recommenders: “XXX is one of the PCI XXX recommenders.”
The authors investigated the origins of androgenetic lineages of Corbicula clams. The origin of a distinct reproductive mode represents a key evolutionary transition, and is of particular relevance in Corbicula because it appears to be associated with increased invasiveness. However, past studies based on one or two markers have failed to pin down the origin of androgenesis. In the present study, the authors analysed sequences of four nuclear markers and COI from several hundred individuals including both invasive androgenetic and native sexual populations. They made use of an analysis of allele pairs in diploids to delineate distinct gene pools and conclude that there are at least three distinct geographic origins of androgenetic lineages. While I think that this study has certainly extended the knowledge of the history of this genus, I have several concerns about core aspects of this study, including the hypotheses tested, conclusions drawn and methodological choices.
Immediately after the above statement of hypotheses, the authors seem to claim that the data support the alternative hypothesis of distinct origins of androgenetic lineages. However, they then immediately state (line 374) “While our results do not disentangle the origin of the peculiar reproductive mode of androgenesis in Corbicula, it shows for the first time the distinct biogeographic origins of androgenetic Corbicula lineages [...]”.
How are the two parts of this statement reconciled? I agree with the first part. Given the extent of hybridisation and ‘nuclear capture’ in androgenetic lineages (as documented in previous studies and this one). It seems impossible to rule out that there was a single origin of androgenesis followed by nuclear captures that could have wiped out traces of the original ancestry of form C/S for example (at least at the four nuclear markers were used in this study). I am therefore confused by the second part of the above quote - the idea that this study supports “distinct biogeographic origins” of androgenetic lineages. If we cannot rule out the hypothesis of a single origin followed by nuclear capture in certain lineages, how can we accept that there are distinct origins?
Perhaps the authors are making a subtle distinction between the (more recent) biogeographic source of a particular invasion and the (possibly older) origin of androgenesis as a reproductive mode. If so, this must be stated more clearly to remove ambiguity. The title might need to change too, as this appears to claim that nuclear captures occurred AFTER distinct origins of androgenetic lineages, rather than nuclear captures simply creating the appearance of distinct origins.
Exclusion of singletons Singleton alleles were excluded as they “are not informative about allele-sharing”. This lead to a huge amount of data being disregarded: nearly 90% of sequences in the case of the atps gene, including all Rlc and Indonesian individuals. Singletons might not be indicative of sharing of identical alleles, but their genetic distances from other alleles are still informative about ancestry. A more quantitative approach that considers all haplotypes and the genetic distances among them would surely add power, and potentially reveal additional sharing of highly-similar (but not identical) haplotypes between sexual and androgenetic lineages. I think it is important to at least show that inclusion of these singleton alleles would support and not contradict the main patterns found using the limited number of shared identical haplotypes.
The results section describing Figure 2 only mentions sharing between C/S and A/R forms at 28S in 3 putatively hybrid individuals (line 251-255). However, Figure S3 (Zone A) suggests allele sharing between a larger number of “FFR3” and “FFR4” individuals. Is it right that there are more than 3 putative hybrids? Perhaps some of these are the C/S individuals mentioned later that share alleles with Lake Biwa at the amy gene (line 279)? Why is this not mentioned in the description of Figure 2? Similarly, the paragraph from line 289-293, describing allele sharing between different invasive lineages does not mention the sharing between C/S and A/R at all. I think omitting these findings from the description of results is problematic as it could create the impression the C/S is more distinct than it really is.
Line 325-326. “In this study, the haploweb approach and the conspecificity matrix gave consistent results for all markers tested.” This statement is misleading. Firstly, the conspecificity matrix is simply a summary of the Haploweb results and does not represent an independent source of information. Secondly, due to the exclusion of singletons, the placement of many individuals in a particular FFR is supported by only one of the four markers, so these individuals cannot be used to support the argument of consistency across markers. Finally, there are several individuals for which the placement is ambiguous (i.e. zones A and B of Figure S3). I therefore think that this part of the paper needs to be reworded to be more true to the findings.
Markers Is any information about the independence of the four nuclear markers available? For example, are they on different linkage groups? This would help with interpretation of cases where the results are consistent among markers. Moreover, I think the paper needs to point out that a larger number of markers, such as GBS or WGS data might be able to resolve the questions further. I appreciate the huge effort that must have gone into generating the sequence data for the present study. However, with hindsight I think it is fairly clear that information from just four markers would be unlikely to resolve the origins of androgenesis given the extent of hybridisation in this genus. It would be a shame for future researchers to invest time in generating similar large data sets from a small number of markers just to discover that they lack sufficient power to infer genomic ancestry. Moreover, assuming that there are loci associated with androgenesis, genome-wide sequence data would potentially enable future researchers to map these loci through association analysis, which could also shed light on its origins.
Samples from previous studies Why were previously studied samples, such as those from Iberia (Peñarrubia et al. 2017) and Russia (Bespalayaet al. 2018) not included in this study? Is it simply because they only used 28S and COI markers? Since so many singletons were excluded from the present study, this doesn’t seem to be a very strong argument for excluding these potentially interesting populations.
Line 342. Since the reproductive mode of the South African individuals was not identified, is it not possible that this represents an invasive population of form C/S rather than a geographic origin?
I personally find Figure S3 to be a more easily interpretable summary of the Haploweb results than the multiple panels of Figure 2. However it would be even more useful if information about the geographic origin or morphological form, or both of each individual can be included along the axes, rather than simply summarising which groups of individuals fall in each clade.
The lines connecting different populations in Figure 3 sometimes differ in width in the two populations. Why is this? I would expect that if 10 alleles are shared between two populations, the line should have width 10 at both ends?
Nuclear capture Fertilization by diploid sperm with retention of maternal chromosomes is called nuclear capture in line 59 and simply hybridization in Figure 1b. Is there a clear distinction, and if not, can a single term be used throughout the paper?
Line 111 cites Doyle 1995 for FFRs, whereas Figure S2 cites Doyle 1992. Is this intentional?
This study investigates the phylogeography of Corbicula clams, a genus that includes both sexual species and asexual relatives. The asexuals have a special reproduction system (“androgenesis”) that results in clonal transmission of the paternal nuclear genome, while the formation of both eggs and sperm are still required. Moreover, due to maternal inheritance of mitochondria, phylogenies may show “cytonuclear mismatch” due to “capture” of mitochondria from divergent lineages. Finally, nuclear maternal chromosomes may sometimes be retained, which potentially leads to formation of hybrid nuclear genomes with increased ploidy and to diversity among asexual lineages.
The study is carried out carefully and the manuscript is well-written. However, the importance of this work beyond the particular study system is not sufficiently well explained. I can see several potential approaches, which may be taken to place the study in a more general framework. For instance, phylogeographic studies in asexuals with complicated reproduction systems may be addressed in a more general way, and the advantages of the main method for data analysis employed here (“allele sharing”) to analyze such systems could be explained more explicitly. The current manuscript, however, is mostly set up from a system-specific perspective. Moreover, several similar studies have been carried out on this system before, and some of the results of the current paper are of confirmatory nature (though being based on a larger sample and more suitable genetic markers than previous studies).
Besides these general remarks on the interest of the manuscript to a general evolutionary readership, I have a few specific comments and suggestions for further improving/clarifying the paper:
This study seeks to better understand the origin of the genetic diversity observed in different asexual lineages of Corbicula clams. The authors investigated the pattern of allele sharing between different asexual lineages and related sexual species using a collection of individuals sampled worldwide. They identify three distinct genetic clusters containing asexual lineages with different biogeographic origins. Generally, I found the approach of using haplowebs to clarify the relationships among the different lineages very interesting. However, I found that some parts of the manuscript were hard to follow, especially for readers that are not familiar with this system. I provide below major and minor comments that I hope will help improve the manuscript and making it more accessible to a general audience.
Major comments:
1/ I found that some basic information regarding androgenesis in this species was missing. This type of information is important as the authors rely on particular assumptions for their haploweb analysis and as this could help interpreting their data. For example, it is unclear whether self-fertilization in androgenetic individuals involves haploid or diploid sperm and whether recombination can occur between homologues (L 42). The authors could also better explain the mode of reproduction of polyploid individuals. For example, in Fig. 1b would the resulting triploid individual produce only haploid gametes with a blue chromosome and diploid gametes with red chromosomes? The authors do not discuss the fact that they do not detect more than two alleles in samples that could potentially be polyploid (L 173). Does that mean that triploids only arise through self-fertilization of diploid or triploid individuals and not through outcrossing with other lineages?
2/ I found that the authors could better explain how recombination between distant haplotypes in sexual or asexual individuals could affect their results. I am not familiar with the haploweb approach, but this seems to be an issue to me. The authors did check for the absence of chimeric sequences and for the validity of their method by performing both cloning and direct sequencing on 5 individuals. However, no information is provided on the way these individuals were chosen. To make a stronger point, I guess I would have chosen individuals from a diverse sexual population such as Lake Biwa, rather than from asexual populations from Europe or America (L177). Also, the authors did not provide the number of clones sequenced per individual. I thought that “(done twice)” meant that they only sequenced two clones per individuals, which seems an unreasonably low number.
3/ I guess the invasive asexual lineages have a very different demography compared to the non-invasive ones. Would it be possible to compare the structure of haplotype networks for these two types of lineages?
L 21: Maybe define “all-male asexuality” as this term is does not really say more than “androgenesis”.
L25-27: These two sentences are not very clear.
L62-119: I find this section hard to follow. Adding a figure with a map would help simplify the text, using a color code similar to the one in Fig. 2 would be useful. I would suggest streamlining this section by listing the different androgenetic lineages and where they are found so that the reader can understand that some forms are invasive (A/R, B, Rlc and C/S) and some others are not (e.g. Vietnam).
L 97: “Nuclear capture between androgenetic and divergent sympatric sexual or androgenetic lineages”?
L126: “form” is a bit obscure, maybe it would possible to be more specific and to mention if these taxa are defined based solely on morphological data or on both morphological and genetic data.
L 163: How would the strong departure from Hardy-Weinberg (high number of heterozygous loci in asexual lineages) affect the phasing?
L 256 “and C. fluminalis africana away from FFR4 individuals?
Fig. 2: I found the captions not very informative. Maybe you could explicitly mention the androgenetic lineages and the sexual species in the caption?
https://doi.org/10.24072/pci.evolbiol.100198.rev14