

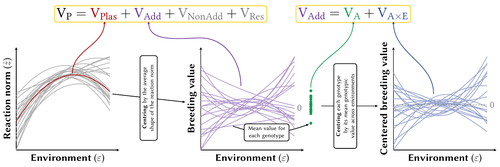
Phenotypic plasticity can lead to rapid and important changes of trait distributions depending on environmental conditions, with important consequences for population dynamics, species interactions and adaptation. To better understand the evolution and importance of plasticity, we need to improve our ability to quantify and compare phenotypic plasticity among organisms, and estimate the evolutionary potential of plastic capacity. Plasticity is classically quantified through regression slopes, with units of traits per environment that by definition vary across organisms, traits and studies, and makes complex the comparison of how does plasticity vary across biological units, especially when linear versus quadratic reaction norms are considered. A clear methodology to quantify phenotypic plasticity in a way that allow for comparison across traits, organisms and environments is lacking.
In this contribution, Pierre de Villemereuil and Luis-Miguel Chevin clarify key concepts about variability and evolutionary potential of plasticity, and propose an efficient method to partition phenotypic variance between genotype and environment. Expanding from the classical (too) simple regression slope approach, and directly integrating the genetic variability of plasticity, they provide a clear framework to quantify the part of phenotypic variance resulting from phenotypic plasticity, integrate the role of the shape of reaction norms, and estimate the heritable variation of trait plasticity. They integrate this method in a R package named Reacnorm, with step by step decision tree, that should greatly help the applicability of this framework.
The authors here propose a contribution that simultaneously clarify key concepts and challenges about plasticity and reaction norms, being very interesting on the theoretical side, and is directly and quite simply applicable to any trait and organism. This should help and stimulate comparative studies of how does plasticity vary across traits, organisms and environmental contexts, both using already published datasets and through new experiments.
References
Pierre de Villemereuil, Luis-Miguel Chevin (2025) Partitioning the phenotypic and genetic variances of reaction norms. EcoEvorxiv, ver.4 peer-reviewed and recommended by PCI Evol Biol https://doi.org/10.32942/X2NC8B
DOI or URL of the preprint: https://doi.org/10.32942/X2NC8B
Version of the preprint: 3
Dear Pierre and Luis-Miguel,
I would like to congrat you for the excellent work you've done for the revised version of your manuscript on partitionning the phenotypic variance of reaction norms. One of the reviewer has raised a few additional minor points that we would like you addressing. Once these final suggestions are incorporated, I believe your manuscript will make a very interesting contribution, combining important theoretical insights with practical applicability, thanks to the clear and accessible tools you've provided.
Thank you again for this contribution. I look forward to read the revised version!
All my best,
Staffan
The authors have responded appropriately to my concerns about the previous version of the manuscript, either through changes in the manuscript or by elaborating on their arguments in their response letter. I still believe that the question adressed by this paper is highly relevent, and I appreciate that the authors have undertaken to create a complete R package for their approach, which I think will make it easier for people interested in plasticity to get to grips with it.
DOI or URL of the preprint: https://doi.org/10.32942/X2NC8B
Version of the preprint: 1
Dear authors,
your manuscript about Partitioning the phenotypic variance of reaction norms has been read by two reviewers. As you will see, they have a number of constructive comments and queries that should be taken into account in a revised version.
Thanks you for your consideration of PCI to publish your work. We look forward to a revised version of your manuscript.
All my best,
Staffan