

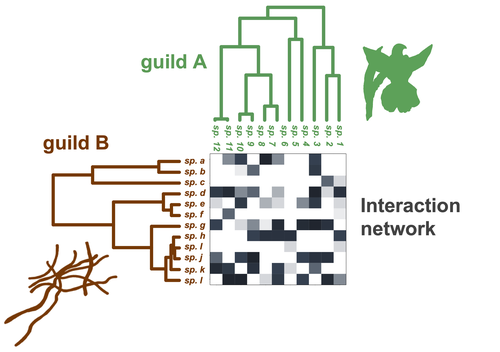
Species are immersed within communities in which they interact mutualistically, as in pollination or seed dispersal, or nonreciprocally, such as in predation or parasitism, with other species and these interactions play a paramount role in shaping biodiversity (Bascompte and Jordano 2013). Researchers have become increasingly interested in the processes that shape these interactions and how these influence community structure and responses to disturbances. Species interactions are often described using bipartite interaction networks and one important question is how the evolutionary history of the species involved influences the network, including whether there is phylogenetic signal in interactions, in other words whether closely related species interact with other closely related species (Bascompte and Jordano 2013, Perez-Lamarque et al. 2022). To address this question different approaches, correlative and model-based, have been developed to test for phylogenetic signal in interactions, although comparative analyses of the performance of these different metrics are lacking. In their article Perez-Lamarque et al. (2022) set out to test the statistical performance of two widely-used methods, Mantel tests and Phylogenetic Bipartite Linear Models (PBLM; Ives and Godfray 2006) using simulations. Phylogenetic signal is measured as the degree to which distance to the nearest common ancestor predicts the observed similarity in trait values among species. In species interaction networks, the data are actually the between-species dissimilarity among interacting species (Perez-Lamarque et al. 2022), and typical approaches to test for phylogenetic signal cannot be used. However, the Mantel test provides a useful means of analyzing the correlation between two distance matrices, the between-species phylogenetic distance and the between-species dissimilarity in interactions. The PBLM approach, on the other hand, assumes that interactions between species are influenced by unobserved traits that evolve along the phylogenies following a given phenotypic evolution model and the parameters of this model are interpreted in terms of phylogenetic signal (Ives and Godfray 2006). Perez-Lamarque et al (2022) found that the model-based PBLM approach has a high type-I error rate, in other words it often detected phylogenetic signal when there was none. The simple Mantel test was found to present a low type-I error rate and moderate statistical power. However, it tended to overestimate the degree to which species interact with dissimilar partners. In addition to the aforementioned analyses, the authors also tested whether the simple Mantel test was able to detect phylogenetic signal in interactions among species within a given clade in the phylogeny, as phylogenetic signal in species interactions may be localized within specific clades. The article concludes with general guidelines for users wishing to test phylogenetic signal in their interaction networks and illustrates them with an example of an orchid-mycorrhizal fungus network from the oceanic island of La Réunion (Martos et al 2012). This broadly accessible article provides a valuable analysis of the performance of tests of phylogenetic signal in interaction networks enabling users to make informed choices of the analytical methods they wish to employ, and provide useful and detailed guidelines. Therefore, the work should be of broad interest to researchers studying species interactions.
References
Bascompte J, Jordano P (2013) Mutualistic Networks. Princeton University Press. https://doi.org/10.1515/9781400848720
Ives AR, Godfray HCJ (2006) Phylogenetic Analysis of Trophic Associations. The American Naturalist, 168, E1–E14. https://doi.org/10.1086/505157
Martos F, Munoz F, Pailler T, Kottke I, Gonneau C, Selosse M-A (2012) The role of epiphytism in architecture and evolutionary constraint within mycorrhizal networks of tropical orchids. Molecular Ecology, 21, 5098–5109. https://doi.org/10.1111/j.1365-294X.2012.05692.x
Perez-Lamarque B, Maliet O, Pichon B, Selosse M-A, Martos F, Morlon H (2022) Do closely related species interact with similar partners? Testing for phylogenetic signal in bipartite interaction networks. bioRxiv, 2021.08.30.458192, ver. 6 peer-reviewed and recommended by Peer Community in Evolutionary Biology. https://doi.org/10.1101/2021.08.30.458192
DOI or URL of the preprint: https://doi.org/10.1101/2021.08.30.458192
Version of the preprint: 4
 , posted 22 Aug 2022
, posted 22 Aug 2022Dear authors,
I have now received comments from the two expert reviewers who previously provided comments on your preprint. Both are happy with how you addressed their previous comments and think the work is almost ready for recommendation. One reviewer raised to minor points which I think could be relatively easily addressed:
• l.162-184: Although the authors added a new figure that helps summarising the simulations (great job!), a table summarising the different parameters could still be useful (I still need to go back to the methods when reading the results part to find which parameter is which).
• l.431: I am still not sure why the authors used a 10 million arbitrary branch length, is this corresponding to some average speciation time in some other studies? Also on Figure 5, these politomies seem younger than 10 Mya.
I would ask you to address these two minor comments and I will make the final decision without sending the work out for review again.
I would end thanking you for considering PCI Evolutionary Biology for your work and also for the positive responses to the reviewer's comments.
Best wishes,
Alejandro
The authors did an excellent job and I do not have further suggestions.
https://doi.org/10.24072/pci.evolbiol.100542.rev21DOI or URL of the preprint: https://doi.org/10.1101/2021.08.30.458192
Version of the preprint: 3
 , posted 16 May 2022
, posted 16 May 2022I have read with interest the submitted preprint Do closely related species interact with similar partners? Testing for phylogenetic signal in bipartite interaction networks. I think the manuscript is well written and clearly presented in general. I think it could make for a valuable contribution to the field given the meticulous analyses of the perfomance of different metrics to test for phylogenetic signal in interaction networks. The two expert reviewers also agree that the work is well written and could be of interest to evolutionary biologists and evolutionary ecologists. The two reviewers have made a number of excellent suggestions on how to improve the work which should be addressed prior to recommendation. I have very little to add to the thorough reviews by both expert reviewers.
I look forward to receiving a revised version of your manuscript.
Perez-Lamarque et al. present a throughout study that tackle most (if not all) common problems when assessing phylogenetic signal in species interactions. They did so by nicely comparing the performance of two commonly used methods (Mantel test and PBLM) on simulated benchmarks. A theoretically more advanced method was also evaluated (PGLMM). The authors considered different scenarios including interaction sign and strength, phylogenetic signal in generalism and sampling asymmetry. Moreover, they developed an interesting procedure to test for phylogenetic signal across different clades. Finally, based on obtained insights, they explored an empirical dataset of orchid-mycorrhizal fungus interactions and aimed at providing general guidelines when measuring phylogenetic signal in ecological interactions. Overall, they found that PBLM (and PGLMM) are more prone to type I errors than Mantel test.
The manuscript is well written and the research is well conducted, timely and provides some interesting findings. I also appreciate the huge effort to conduct the large battery of simulations and analyses while keeping a congruent manuscript. I have, nevertheless, some conceptual and methodological considerations that may strengthen the research. It must be noted that I am more familiar with the Mantel test and my suggestions are focused in that direction.
1. Comparing Jaccard vs UniFrac distances
The authors compared the performance of Jaccard and UniFrac distances to detect phylogenetic signal in interaction partner use. They found that UniFrac distances outperformed Jaccard distances and concluded: “we advocate the use of weighted UniFrac distances” (line 689). The point here is that Jaccard and UniFrac distances are measuring dissimilarities in different aspects of interacting species (taxonomic vs phylogenetic compositional dissimilarities, respectively) that may reflect different evolutionary processes. For instance, assuming that interactions are trait-mediated and following the author’s nomenclature, if the traits that regulate interactions are conserved in guild A but not in guild B, then we should find that phylogenetically related species of guild A would share interaction partners of guild B that are unrelated. Thus, for guild A, we could expect phylogenetic signal in species interactions when using Jaccard distances but not when using UniFrac distances (perhaps Fig. 1A in Calatayud et al. 2016 might help, and sorry for the self-advertising). This exemplified that Jaccard and UniFrac distances (either weighted or not) can reflect different processes and, therefore, that they cannot be safely compared. I would suggest to remove the direct comparison and subsequent recommendations between these two distance indices.
2. Effects of the number of interacting partners (and other generalism levels)
The authors proposed a sequential Mantel test to overcome confounding effects of phylogenetic signal in the number of interacting partners. They conducted a first Mantel test to explore the correlation between phylogenetic and ecological distances and a second one to explore the correlation between phylogenetic distances and differences in the number of interacting species. If both correlations were significant they treated the former as non-significant. While I agree that this approach may work in some situations, it will certainly produce type II errors when both composition and number of partners show phylogenetic signal, as they recognize in the discussion.
I think there are better alternatives to solve this issue than the sequential Mantel test. The root of the problem here is that both the Jaccard and the UniFrac distances take into account differences in taxonomic (i.e. number of species) and phylogenetic (i.e. sum of phylogenetic branch lengths) generalism, respectively. Hence, species with different levels of taxonomic or phylogenetic generalism will also show differences in interacting partner use when using these indices. This is a common issue in many other situations, and there are well-stablished dissimilarity indices to overcome it (Baselga 2010, Leprieur et al. 2012, see also Calatayud et al. 2016 for their use in a similar context). By only taking into account dissimilarities due to true changes in the partner species/phylogenetic composition, these indices are robust to produce spurious correlations between ecological and phylogenetic distances when generalism levels show phylogenetic signal.
Alternatively (or even better complementarily), using appropriate randomization schedules to asses statistical significance in Mantel test can help to get rid of this confounding effect. That is, rather than permuting any of the distance matrices (as I guess the authors did), one can permute the raw interaction matrix by retaining some of its properties. For this case, it would be possible to randomise the interaction matrix keeping constant the number of interaction partners. While this does not affect observed correlation coefficients, it certainly reduces type I errors associated with phylogenetic signal in generalism levels, improving also other issues of Mantel test (Guillot & Rousset 2013). Note also, that randomizations of raw data can also accommodate other aspects such as unequal sampling effort or spatial patterns (e.g. Vázquez et al. 2009), making Mantel test highly flexible (perhaps this could also be discussed).
In summary, by using appropriate dissimilarity indices and null models it is possible to remove the effects of potential phylogenetic signals in generalism levels. To the best of my knowledge, this is where the state of the art is when using the Mantel test (or any of its updates, see, for example, Ferrier et al. 2007 for generalized regression on distance matrices). Still, the behaviour of these apparently and theoretically more advanced approaches has not been tested using simulated benchmarks. I think the authors have the perfect opportunity to do this, which I believe would certainly improve the research. Though I encourage the author to test this approach, I am totally aware that it might imply a huge (perhaps unfeasible) effort. If this is the case, I would suggest to send the sections “Confounding effect of the phylogenetic signal in the number of partners” to supporting information, especially considering that your simulations should not produce phylogenetic signal in generalism. At the very least, I think the commented alternatives deserve a mention in the discussion, perhaps removing any recommendation to the sequential Mantel test.
3. Minor considerations
I am just not sure whether the mantel test is not also a model-based approach as it implicitly assumes that ecological divergence increase with divergence time (Letten & Cornwell 2015). Indeed, phylogenetic distances used in Mantel test (or analogous) can be modified to accommodate different evolutionary models (e.g. Calatayud et al. 2019). Of course, it is not as explicitly as other models, but I think this classification might be controversial. Just not sure.
Figure 2. I think it should be “Guild A” instead of “Clade A”.
Other potential missing references for the introduction:
Braga, M. P., Janz, N., Nylin, S., Ronquist, F., & Landis, M. J. (2021). Phylogenetic reconstruction of ancestral ecological networks through time for pierid butterflies and their host plants. Ecology Letters, 24(10), 2134-2145.
Futuyma, D. J., & Agrawal, A. A. (2009). Macroevolution and the biological diversity of plants and herbivores. Proceedings of the National Academy of Sciences, 106(43), 18054-18061.
Gómez, J. M., Verdú, M., & Perfectti, F. (2010). Ecological interactions are evolutionarily conserved across the entire tree of life. Nature, 465(7300), 918-921.
4. References
Baselga, A. 2010. Partitioning the turnover and nestedness components of beta diversity. Global Ecology and Biogeography 19:134-143
Calatayud, J., Hórreo, J. L., Madrigal-Gonzalez, J., Migeon, A., Rodriguez, M. A., Magalhaes, S., & Hortal, J. (2016). Geography and major host evolutionary transitions shape the resource use of plant parasites. Proceedings of the National Academy of Sciences, 113(35), 9840-9845.
Calatayud, J., Rodríguez, M. Á., Molina-Venegas, R., Leo, M., Horreo, J. L., & Hortal, J. (2019). Pleistocene climate change and the formation of regional species pools. Proceedings of the Royal Society B, 286(1905), 20190291.
S Ferrier, G Manion, J Elith, K Richardson, Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Divers Distrib 13, 252–264 (2007).
Guillot, G., & Rousset, F. (2013). Dismantling the Mantel tests. Methods in Ecology and Evolution, 4(4), 336-344.
Leprieur F, Albouy C, De Bortoli J, Cowman PF, Bellwood DR, et al. (2012) Quantifying Phylogenetic Beta Diversity: Distinguishing between "True" Turnover of Lineages and Phylogenetic Diversity Gradients. PLoS ONE 7(8): e42760.
Letten, A. D., & Cornwell, W. K. (2015). Trees, branches and (square) roots: why evolutionary relatedness is not linearly related to functional distance. Methods in Ecology and Evolution, 6(4), 439-444.
Vázquez, D. P., Chacoff, N. P., & Cagnolo, L. (2009). Evaluating multiple determinants of the structure of plant–animal mutualistic networks. Ecology, 90(8), 2039-2046.
https://doi.org/10.24072/pci.evolbiol.100542.rev11