

 based on reviews by Miguel de Navascués and 1 anonymous reviewer
based on reviews by Miguel de Navascués and 1 anonymous reviewer
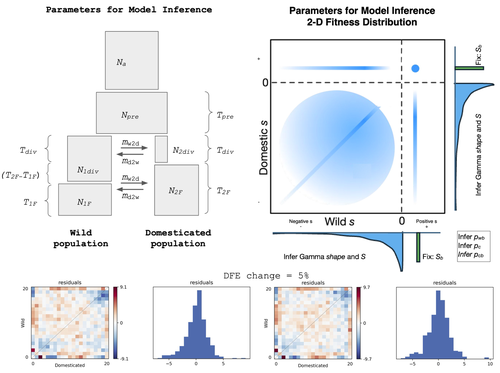
The joint full distribution of fitness effects (DEF) is an important indicator in population genetic studies, and its inference has been the subject of intense research [1]. However, we still lack a solid framework to estimate DFE under certain demographic conditions.
In this study, Castellano and colleagues propose to estimate the DFE by analysing the site frequency spectrum (SFS), and specifically develop a new approach for the joint DFE model inference [2]. The latter is based on the proportion of variants with divergent selection coefficients. Authors performed extensive simulations under models of domestication which is arguably one of the most crucial series of events in human evolution [3]. Domestication is associated with significant genetic costs in animals [4].
While DFE is typically estimated by contrasting SFS of silent and functional mutations [5], it has been recently suggested to use the joint SFS between domesticated and wild populations to estimate the DFE [6]. Authors build on this model and expand its parameterisation. Authors were able to dissect the impact of linked selection on inferred demographic history of wild and domesticated populations, with a robust estimation of the deleterious DFE.
There are still several limitations in the interpretation of DFE as, for instance, some selective sweeps can bias their estimates and some demographic scenarios are challenging to infer. Also, classic quantitative trait models should be evaluated as a complementary approach. Finally, the in silico predictions presented in this study could be validated by empirical scans on existing genomic data sets. Nevertheless, this study is an important contribution to our understanding on how demography, and domestication in particular, can affect variants under selection in recent evolutionary histories.
References
[1] Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8(8):610-618. https://doi.org/10.1038/nrg2146
[2] Castellano D, Vourlaki IT, Gutenkunst RN, Ramos-Onsins SE. Detection of Domestication Signals through the Analysis of the Full Distribution of Fitness Effects. BioRxiv 2025, ver.4 peer-reviewed and recommended by PCI Evol Biol https://doi.org/10.1101/2022.08.24.505198
[3] Frantz LAF, Bradley DG, Larson G, Orlando L. Animal domestication in the era of ancient genomics. Nat Rev Genet. 2020;21(8):449-460. https://doi.org/10.1038/s41576-020-0225-0
[4] Schubert M, Jónsson H, Chang D, et al. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc Natl Acad Sci U S A. 2014;111(52):E5661-E5669. https://doi.org/10.1073/pnas.1416991111
[5] Kousathanas A, Keightley PD. A comparison of models to infer the distribution of fitness effects of new mutations. Genetics. 2013;193(4):1197-1208. https://doi.org/10.1534/genetics.112.148023
[6] Huang X, Fortier AL, Coffman AJ, et al. Inferring Genome-Wide Correlations of Mutation Fitness Effects between Populations. Mol Biol Evol. 2021;38(10):4588-4602. https://doi.org/10.1093/molbev/msab162
 , 27 Jan 2025
, 27 Jan 2025The authors have thoroughly revised their manuscript in response to the suggestions from the previous review. The new version is significantly clearer, and the additional results complement the earlier work. I recommend the publication of this manuscript.
DOI or URL of the preprint: https://doi.org/10.1101/2022.08.24.505198
Version of the preprint: 3
 , posted 25 Jun 2024, validated 25 Jun 2024
, posted 25 Jun 2024, validated 25 Jun 2024Dear authors,
thank you for your submission. Your study has been evaluated by two experts in the field. Both of them find it of relevance and importance, as your results provide a good contribution to the field. The focus on domestication has also been appreciated.
However, reviewers point to several concerns with your submission. Both indicate a lack of details in the description of the methods, especially with the new approach to infer demography and selection. The code provided on github is not sufficiently documented. There are also some concerns over the interpretation of some results, where the reviewers appear to be less optimistic than you on the performance of your inferences. I also find Figure 3 to be too cluttered.
While the study has merit, I encourage the authors to carefully consider all the points raised by the reviewers to clarify your text accordingly.
Matteo
Title and abstract
Does the title clearly reflect the content of the article? Yes
Does the abstract present the main findings of the study? Yes
Introduction
Are the research questions/hypotheses/predictions clearly presented? Yes
Does the introduction build on relevant research in the field?Yes, but it lacks some citations. I have added those to my review.
Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? No (please explain) The methods are not presented in detail. I have specified that in my review.
Are the methods and statistical analyses appropriate and well described? No (please explain)
Results
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? I don’t know
Are the results described and interpreted correctly? Yes
Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? Yes
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? Yes
 , 14 Jun 2024
, 14 Jun 2024Title and abstract
Introduction
Materials and methods
Results
Discussion
In this work, the authors address the performance of statistical methods to infer demography and selection (distribution of fitness effects, DFE) under an evolutionary scenario representing a domestication process. Some of the methods evaluated are widely used in the community, and one is a newly developed approach that accounts for the change of selection regime in the domesticated population. While the results reflect previously published evaluations of the negative effects of linked selection on demographic inference and the challenges of disentangling selection and demographic effects, the work merits attention for its specific focus on domestication and the development of a new approach. Contrary to the opinion of previous reviewers, I argue that the domestication scenario is a societally important evolutionary process with broad interest. Moreover, a scenario of divergent populations with one undergoing a bottleneck and a change of selection regime may also be relevant in other contexts, such as the case of invasive species. Unfortunately, the text presenting the work remains unclear, particularly regarding the description of the new approach. Below, I provide a list of points (roughly ordered from "concerns" to "suggestions") that need to be addressed before the work can be recommended.