

 based on reviews by Qi Zhou and 2 anonymous reviewers
based on reviews by Qi Zhou and 2 anonymous reviewers
There is a heavy body of literature on sex-biased gene expression, which can easily be tricky. One reason is that expression data are multi-dimensional, notoriously noisy, and highly sensitive to experimental conditions. Achieving reproducibility is, therefore, a challenge, especially in non-model organisms. Another reason is that the evolutionary forces shaping gene expression variation are complex, involving processes such as intra- and inter-locus conflicts between sexes, sex-chromosome evolution and degeneration, dosage compensation, cis vs. trans regulation, dominance, linkage, drift, etc... Not surprisingly, the field is rich in discordant studies, and a wide variety of situations have been described.
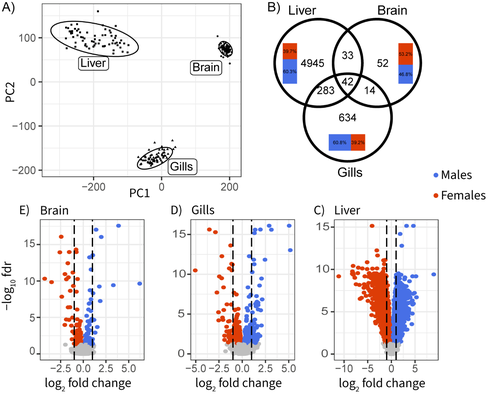
The three-spined stickleback is a good illustration. This widely-studied fish displays conspicuous sexual dimorphisms, both morphological and behavioural. Yet, the existing literature regarding which genes and tissues are involved is remarkably unclear due to the lack of a comprehensive data set. Sylvestre et al. (1) here fix this issue. They sampled 40 wild-caught individuals in each of the two sexes and performed high-throughput sequencing of the transcriptome in three somatic tissues, dissected and preserved in the exact same conditions. This is an impressive effort, well above current standards. Data analysis delivered a series of neat results: gene expression in the liver is particularly sex-biased; the brain, in contrast, is remarkably little sex-differentiated despite the presence of courtship and paternal care in this species; gills show significantly sex-biased gene expression, which had been unnoticed previously despite the importance of this organ in fish ecotoxicology; the relatively young sex-chromosomes, finally, do not seem to experience dosage compensation, and are therefore enriched in sex-biased genes.
Some of these results are consistent with previous studies in other fish species (2), here confirmed or demonstrated with a high degree of certainty. Others are new and worth considering in future studies of stickleback ecology and reproduction. We simply need more studies of this sort: well-conducted and clear-cut, recalling the career of its last author.
References
(1) Florent Sylvestre, Nadia Aubin-Horth, Louis Bernatchez (2024) Sex-biased gene expression across tissues reveals unexpected differentiation in the gills of the threespine stickleback. bioRxiv, ver.2 peer-reviewed and recommended by PCI Evol Biol https://doi.org/10.1101/2024.06.09.597944
(2) Iulia Darolti I, Judith E. Mank (2023). Sex-biased gene expression at single-cell resolution: cause and consequence of sexual dimorphism. Evolution Letters 7(3):148-156. https://doi.org/10.1093/evlett/qrad013
DOI or URL of the preprint: https://doi.org/10.1101/2024.06.09.597944
Version of the preprint: 1
 , posted 04 Sep 2024, validated 04 Sep 2024
, posted 04 Sep 2024, validated 04 Sep 2024Dear authors,
Thanks for submitting this preprint to the PCI Evol Biol. I found the manuscript attractive due to the general interest of the topic (sex-biased gene expression, sex chromosome evolution) and the quality of the data you gathered. Three colleagues have now reviewed your work. They all agree about the strengths of the study, while also consistently asking for a more elaborate discussion of its implications, in the light of the existing literature. I find the reviews quite positive and useful. I suggest addressing all the comments in a revised version which, I anticipate, should make a valuable contribution to the literature.
Please find below a couple of additional line-by-line comments by myself. Some of these are redundant with the reviewers' as they were made independently.
Looking forward to reading the revision, best regards,
Nicolas Galtier, CNRS - University Montpellier
---------
Nicolas Galtier's comments
- l41: "Sex chromosome thus": the logical link between this sentence and the previous one does not appear obvious
- l51-57: how meaningful is the comparison of numbers of detected sex-biased genes across distinct studies? do they share the same approaches, power, thresholds, etc...?
- l67: I would suggest saying: "The lowered effective size of the Y (or W) chromosome, present in only one sex and therefore non-recombining, …" as this is the main point of the cited Charlesworth & Charlesworth article.
- l182: reference to "(lien?)" to be removed/modified
- l83-97: I feel like a better job could be made connecting this bibliographic survey to your study. What questions are left open following the reviewed studies, which you would like to address? Are there limitations/heterogeneity across studies (sample size, methodology, data analysis) that need to be overcome? You mention the use of lab-raised individuals, which is fine; could you perhaps elaborate on why your study is warranted and what novel interesting knowledge is expected to be produced, given the existing literature?
- A related comment would be that the discussion hardly compares your results to the existing ones, or addresses the reasons for the discrepancies, when there are some.
- l106: shouldn't the reference to Kotrschal et al. 2012 be part of the previous paragraph? (l83-91). This sounds like an important piece of the state of the art, but it is disconnected from the rest of the bibliographic survey.
- l166: you mention using a method that differs from the classical negative binomial model, but no description of the method is made and no reference is given, besides the generic Benjamini-Hochberg paper. Is the method described in the DESEQ2 manual? If yes please state it, maybe refer to the manual section, and maybe add a couple of words describing it.
- l332: "Synaptic signaling..." -> this sentence seems to say the same thing twice?
- l337: "This suggests that gene ontology analysis in gills suffer from the lack of gill-specific information." I'm not sure what exactly this sentence is intended to mean, but it relates to a comment I'm having on this and the next (liver) section. GO enrichment analysis was here performed by comparing the annotations associated to DEGs in gills to the whole set of annotations associated to the three-spine stickleback genome. This is fine but we're left with the following question: are gill sex-biased genes enriched in ion-related and immunity genes because they are sex-biased, or because they are gill-expressed? This could be examined by contrasting annotations between gill DEGs and all gill-expressed genes, or between all gill-expressed genes and whole genome (and same for liver).
- Not being a fish expert I was not aware that extensive sex-biased gene expression in the liver is documented in this group. Your results strongly corroborate this previous finding. Maybe would you like to recall what is known or thought about the reasons for this pattern? Why the liver and not the brain, for instance? Is this specific to fishes? Feel free to comment on others' suggestions if any, and to share your own hypotheses.
Sylvestre et al. describe a fantastic dataset for exploring sex-specific patterns of expression in wild-caught sticklebacks, with over 30 individuals of each sex sequenced for RNA of brain, gill and liver. They describe substantial heterogeneity between tissues in the amount and functional nature of sex-biased genes in the three tissues, and confirm that dosage compensation has not evolved in this group in response to Y chromosome degeneration.
I have some questions below about data acquisition and analysis, which I think are worth clarifying in the manuscript.
* In response to the specific questions asked by PCI: I found the title a bit disconnected from the article, since gills are a small part of the results, and most of the sex-bias is found in the liver. But that's a matter of personal taste, I also understand why you would chose to emphasize the most surprising result.
* "We collected adult anadromous three-spined sticklebacks from tide pools of the St Lawrence
120 River at Baie de l’Isle verte (48.009961, −69.407070). "
--> Do you have a sense of how old the individuals were? Would age heterogeneity affect the results?
* "In fish, global dosage compensation has rarely been found (Darolti et al., 2019) and
81 we still lack knowledge about the extent of the evolution of dosage compensation in this highly
82 diverse group."
--> this sounds slightly confusing, as it could mean that global compensation has not been found in fish with differentiated sex chromosomes, or that it has not been studied because there are not so many fish species with differentiated sex chromosomes. Could you clarify?
* "Identification of Shared Genes Between X and Y"
--> I could not understand how this was done. How exactly do the custom scripts work? What information is used to know if you have X and/or Y-derived transcripts? Are X- and Y-linked genes already annotated? Could you make the methods more explicit?
* "We
246 had a percent mapping of uniquely mapped reads to the reference genome ranging from 44.3% to
247 88.61% (median 76.76%)."
--> This seems low. Could you provide numbers for each sample (perhaps this is already in a supplementary table, I could not find these), and perhaps repeat analyses with only samples with good mapping rates as an additional line of evidence that the conclusions are robust? More importantly, are there systematic differences in mapping rates between tissues? These could affect your power to detect sex-bias.
* On a similar note, while the PCA is reassuring, could you also provide correlation values between replicates? Or more generally a measure of heterogeneity within the sexes as well as between them? From the PCA it seems that liver is highly heterogeneous independent of sex, brain is highly homogeneous, and gills are somewhere in between, so sex-bias may just mirror general patterns of heterogeneity.
* Along those lines, would it be possible to color males and females in the PCA?
* Do you have any kind of mininum expression filtering? If not it could be reassuring to see that patterns hold when very low expression genes are removed, or you could justify why this was not deemed useful.
* "After filtering, we identified five overexpressed genes in the liver but none
248 in the gills or the brain."
--> overexpressed relative to what? Could you define this more specifically?
* 418 Sex-biased Gene Expression on Sexual Chromosome Mostly Reflects Gene Loss in Non-
419 Recombining Regions
It would be helpful to know how many genes are being considered in each of the categories (diploid, hemiploid X, hemiploid Y)
* Fig 5: Why do you have all and diploid in the PAR? Should all PAR genes not be diploid?
Review of the manuscript «Sex-biased gene expression across tissues reveals unexpected differentiation in the gills of the threespine stickleback»
In the manuscript untitled «Sex-biased gene expression across tissues reveals unexpected differentiation in the gills of the threespine stickleback», the authors explore the patterns of sex-biased gene expression in brain, liver and gills during the reproductive period in males and females of the sexually dimorphic threespine stickleback. Confirming previous results, they found that expression in the liver is extremely sex-biased, whereas the brain is not, and that there is a lack of global dosage compensation in this young sex chromosome system. They also found that gills exhibit high levels of differentiation, an interesting result as sex is rarely considered in physiological and ecotoxicological studies of gill responses in fishes. They outline a few genes and gene functions that are particularly sex-biased, and thus likely determinant in sexual dimorphism in this species. Those descriptive results bring some insights into the role of sex biased expression in sexual dimorphism in sticklebacks. It would be even more apparent if the authors added a brief description, for instance in the introduction, of the sexually dimorphic phenotypic traits observed in the focal species for the non-specialist reader. The manuscript is well written, the topic is well introduced, the used methodology is sound (notably the choice of sampling during the reproduction period, and to use the QuanSeq 3’ UTR technology for sequencing) and the analyses are clear and well conducted (only a few details of the material and method require some clarification) and the conclusions adequately supported by the results.
Comments:
1. A non-specialist reader would greatly benefit from a brief description of the phenotypic traits that are under sexual dimorphism in the threespine stickleback (for instance in the introduction at around line 84). It would also be interesting to link the gene ontology observations to those phenotypic traits. For instance, you found several GO enrichment terms related to immune system in SBG, are there any indication that male and female show different immune response in some context?
2. I really appreciated that the a priori expectations were clearly stated at the end of the introduction. It would be even better if they were more deeply discussed later in the manuscript: for instance the expectation that the brain would be highly sex-biased during the reproduction period (line 106-107) is eventually not met in the results : what could be some possible explanations?
3. Maybe briefly explain the principle and advantages of the use of the QuanSeq 3’ UTR technology to quantify gene expression level, and how this might impact comparisons with other studies based on classic RNAseq.
4. In the “Alignment and expression counts” section of the material and method:
-What HTSeq parameters/functions have been used ?
-Multi-mapping reads have to be filtered out to obtain reliable read counts. However, for X-linked genes still retaining a Y copy outside the PAR, I expect reads could map on both the X and Y copy in males. How was this handled for the quantification of expression of XY gametologs? A related question: how was X and Y allele expression measured for those gametologs?
5. For the non-specialists, briefly explain what the different normalization methods in different analyzes normalize for (within sample/within dataset/between samples comparisons) : cpm, Vst (it should be stated that it stands for Variance stabilizing transformation), and the the (average? Median?) ratio normalization factor.
6. Foie/ Cerveau / Branchie are kept in french in supplementary table 1 and 2.
7. At line 121: under five, seven and ten minutes respectively → after death?
8. At line 119: What year were the samples collected?
9. At line 122: We disrupted samples in
10. At line 182: (lien ?) was kept in the text → https://zenodo.org/records/11477976
11. At line 312: (0.78%) of expressed genes) → (0.78% of expressed genes)
12. At line 361: Is it chromosome V, as stated in the text, or chromosome VII, as stated in Figure 2 legend, which is enriched in male-biased genes?
13. At line 366: total gene expression
14. At line 429-431: Precise the statistics that are stated there.
15. At line 450-453: “In sticklebacks, hemizigous genes tend to be dosage insensitive, meaning that protein quantities are independent from its expression level (Peichel et al., 2020).” → I would alleviate this sentence, as the Peichel et al., 2020’ result only indicate that genes with a retained X and Y copy were more likely to exhibit haploinsufficiency based on orthology with human genes.
“This suggests that there is no selective pressure to evolve dosage compensation and is corroborated by the fact that conserved genes are dosage-sensitive and evolving under purifying selection (White et al., 2015).” → I am not sure I understand the second part of this sentence: is it that as Y-linked genes show signs of degeneration, they are likely to be under relaxed selective pressure and thus to be dosage-insensitive?
16. At line 477-484: (p-value=2.10-2)
Sex biased genes are widely found throughout the animal kingdom as potentially the result of resolved sexual conflict. This work inspected the pattern of sex biased genes in gill, brain and liver from the wild population of threespine sticklebacks, a model species for teleost sex chromosome evolution. The species has been previously reported to evolved three recombination suppression events (evolutionary strata) between the XY chromosomes. This work found liver harbors many more sex biased genes than the other tissues, with also many sex biased genes unexpectedly in the gill. Overall I found the work interesting and the conclusions are generally consistent with what one would expect or what the previous results reported. Since there are several works (Kitano et al. 2010, Leder et al. 2010, Kaitetzidou et al. 2022) did a similar study but with different techiniques, or sample sources, I suggest deeper comparison with them, particulary in the regard of lab vs. wild collected samples, as stated in the introduction by the authors. Other aspects that need to be addressed is the techinical aspect of the study, including the definition of sex biased genes, that could lead to very different conclusions. For example, the authors first stated over 5000 sex biased genes in the liver, but then it was reduced to around 300 if the authors applied the same cutoff with one of the previous study, if I understand it correctly. Another contradiction is the feminization pattern in the PAR, that the author found a different result compared to a previous study, due to the usage of autosome or outgroup orthologs, but which one is more appropriate? Finally, a general issue is that the paper is organized in a way that the authors reported the number or general patterns of sex biased genes, then decribe invidiual genes in these tissues. Somehow the reader cannot have a coherent conclusion from these pattern throughout the paper. For example, why gill, a tissue that is not expect to have many sex biased genes show so many in this study? This has not been discussed before and newly found in this study. The author might look deeper into this, just to draw a difference from all previous studies. I have many detailed commments that I marked in the pdf uploaded together with these comments.
Download the review