

 based on reviews by Oscar Lao Grueso and 2 anonymous reviewers
based on reviews by Oscar Lao Grueso and 2 anonymous reviewers
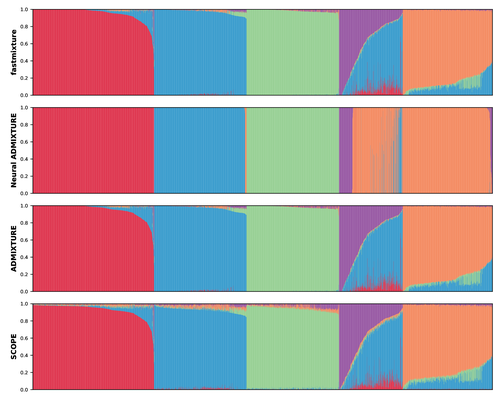
The estimation of ancestry proportions in individuals is an important analysis in both evolutionary biology and medical genetics. However, popular tools like ADMIXTURE (Alexander et al. 2009) and STRUCTURE (Pritchard et al. 2000) do not scale well with the large amount of data currently available. Recent alternative methods, such as SCOPE (Chiu et al. 2022), favour scalability over accuracy.
In this study, Santander and coworkers introduce a new software, called fastmixture, which estimates ancestry proportions and ancestral allele frequencies using novel implementations for initialisation and convergence of its model-based algorithm (Santander et al. 2024). In simulated datasets, fastmixture displays desirable properties of speed and accuracy, with its performance surpassing commonly used software (Alexander et al. 2009, Pritchard et al. 2000, Chiu et al. 2022, Mantes et al. 2023). fastmixture is almost 30 times faster than ADMIXTURE under a complex model with five ancestral populations, while retaining similar accuracy levels. When applied to data from the 1000 Genomes Project (1000 Genomes Project Consortium 2025), fastmixture recapitulated expected levels of global ancestry. The new software is freely available on GitHub with an accessible documentation. fastmixture accepts input files in PLINK format.
It remains an open question whether extensive parameter tuning could increase the scalability and accuracy of established methods. A comprehensive assessment of fastmixture over a wide range of data processing options (Hemstrom et al. 2024) is also missing. Finally, whether model-based approaches are fully scalable to ever increasing biobank datasets is still under debate. Nevertheless, the superior computational performance of fastmixture is evident and it is likely that this new software will soon replace existing popular tools to estimate global ancestry proportions.
References
Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655-1664. https://doi.org/10.1101/gr.094052.109
Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945-959. https://doi.org/10.1093/genetics/155.2.945
Chiu AM, Molloy EK, Tan Z, Talwalkar A, Sankararaman S. Inferring population structure in biobank-scale genomic data. Am J Hum Genet. 2022;109(4):727-737. https://doi.org/10.1016/j.ajhg.2022.02.015
Santander CG, Refoyo Martinez A, Meisner J. Faster model-based estimation of ancestry proportions. bioRxiv 2024; ver.2 peer-reviewed and recommended by PCI Evol Biol. https://doi.org/10.1101/2024.07.08.602454
Mantes AD, Montserrat DM, Bustamante CD, Giró-I-Nieto X, Ioannidis AG. Neural ADMIXTURE for rapid genomic clustering. Nat Comput Sci. 2023;3(7):621-629. https://doi.org/10.1038/s43588-023-00482-7
1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68-74. https://doi.org/10.1038/nature15393
Hemstrom W, Grummer JA, Luikart G, Christie MR. Next-generation data filtering in the genomics era. Nat Rev Genet. 2024;25(11):750-767. https://doi.org/10.1038/s41576-024-00738-6
DOI or URL of the preprint: https://doi.org/10.1101/2024.07.08.602454
Version of the preprint: 1
 , posted 24 Sep 2024, validated 25 Sep 2024
, posted 24 Sep 2024, validated 25 Sep 2024Your submission has now been reviewed by three experts in the field. They are all positive about your study but raise important points.
While it is notable that the new implementation is supposidely faster, an assessment of which improvement is most significant would be of interest. More importantly, the comparison against competing methods could involve more complex scenarios to really appreciate the potential novel contribution of fastMixture to the field. The github repository must include clear information on the version control requirements and a toy example to run.
These changes are essential to prove that fastMixture is going to replace Admixture in the future, as stated in your study.
Please ensure that you address all the points raised by the reviewers or justify why those changes are not needed or outside of the scope of the study.
Review for Manuscript "Faster Model-Based Estimation of Ancestry Proportions"
The authors introduce a software tool, fastmixture, which infers ancestry proportions and allele frequencies within the same likelihood framework used by the frequently used ADMIXTURE software. They propose three novel computational enhancements to speed up the analysis of large datasets. These improvements include:
- An SqS3 acceleration scheme for the EM algorithm,
- A randomized singular value decomposition (SVD) for better initialization of allele frequencies and ancestry proportions,
- Mini-batch updates for the EM algorithm.
Although these improvements do not reduce the computational complexity, the authors state that together, they result in a 20-fold speedup. (Could not check this yet, see comment 5 below)
Main Comments:
1.) It would be valuable to understand which of the three improvements contributes most to the performance gains. If the authors could provide details on the individual impact of each enhancement, this would add useful context.
2.) The performance improvement is substantial and could significantly enhance the workflow of many large-scale genomic studies, without requiring the adoption of an entirely new and potentially less comparable modeling framework. However, for fastmixture to become a viable replacement for ADMIXTURE in future studies, additional tests and direct comparisons with ADMIXTURE would be beneficial.
For instance, I am curious whether the SVD initialization step affects the number of modes (see https://doi.org/10.1093/bioinformatics/btw327) inferred by FastMixture in comparison to ADMIXTURE.
Additionally, over- or under-specifying the number of populations, K, might affect fastmixture differently than ADMIXTURE.
3.) The simulated scenarios appear to be fairly narrow, and expanding the range of population structures could provide more insights. For example, all the scenarios presented (Figures S1 and S2) involve just one admixed population, with variations only in the number of non-admixed populations. There seems to be no ongoing migration between simulated populations. It would be interesting to see whether FastMixture performs similarly to ADMIXTURE in more complex population histories, such as those with multiple admixed populations or constant migration.
4.) I am also curious why NeuralAdmixture performs poorest among the evaluated methods. In the NeuralAdmixture paper, performance did not seem to drop significantly for admixed populations. Perhaps the authors could provide more insights here.
5.) The tool on GitHub was easy to install. However, the script produced an error when we ran it on our example files. This could be due to issues with the local package versions, but currently, there is no way to verify this, as I couldn't find clear version requirements in the github repository. It would be helpful if the GitHub repository included explicit requirements and a minimal working example to make installation verification easier.
The Manuscript:
The manuscript is well-organized, with a clear explanation of the motivation behind the research and the methods employed. I found it easy to place the manuscript within the broader context of related work. Other than the lack of detail on the impact of individual improvements, the manuscript does a good job explaining the concepts. A short paragraph on the principles behind SVD, similar to the description of the SqS3 algorithm, could make the ideas more accessible to readers.
Apart from this, I found the manuscript easy to follow and enjoyed reading it.
Further Comments Regarding the Figures:
Figure 1: Sorting individuals within each subpopulation by the ancestry proportions inferred by fastmixture could help make the distribution of ancestry proportions in the admixed population more visually clear. This suggestion applies to the other figures as well, especially Figure 2.
Figure 3: This would be more effective as a table.
Does the title clearly reflect the content of the article? [X] Yes, [ ] No (please explain), [ ] I don't know
Does the abstract present the main findings of the study? [X] Yes, [ ] No (please explain), [ ] I don’t know
Are the research questions/hypotheses/predictions clearly presented? [X] Yes, [ ] No (please explain), [ ] I don’t know
Does the introduction build on relevant research in the field? [X] Yes, [ ] No (please explain), [ ] I don’t know
Are the methods and analyses sufficiently detailed to allow replication by other researchers? [X] Yes, [ ] No (please explain), [ ] I don’t know
Are the methods and statistical analyses appropriate and well described? [ ] Yes, [X] to some extent, [ ] No (please explain), [ ] I don’t know
Are the results described and interpreted correctly? [X] Yes, [ ] No (please explain), [ ] I don’t know
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? [X] Yes, [ ] No (please explain), [ ] I don’t know
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? [X] Yes, [ ] No (please explain), [ ] I don’t know
The method proposed in this study combines various machine learning and optimization algorithms to significantly reduce the time required for estimating ancestry proportions while producing cutting edge results. The article presents an innovative approach to minimizing computation time by integrating different techniques from the field of machine learning, which I found very insightful and enjoyable to read. I agree with the authors that this methodology has the potential to "be the preferred alternative to ADMIXTURE in future population genetic studies".
However, my main concern lies in how the proposed methodology compares with other existing algorithms. Many established methods assume marker independence and unrelated individuals (for instance, see Methods Mol Biol. 2020; 2090: 67–86. doi:10.1007/978-1-0716-0199-0_4). In contrast, based on the simulation study, it appears that only markers with a minor allele frequency (MAF) below 0.05 are excluded from the analysis. Even when the authors use a subset of markers from the 1000 Genomes Project, this subset is selected randomly. I believe that some of the discrepancies observed between methods could stem from this bias.
Additionally, the simulated models considered in the study raise some concerns. While these models are relevant for studying human demography, they seem rather specific. It would be beneficial to include other, more complex models, especially since the authors assert that 'Our findings suggest that the added noise in the ancestry proportions estimated in SCOPE will only increase for scenarios with a larger K and for more complex demographic models.'"
I have some additional comments and questions regarding the tests conducted:
Please provide references for the values mentioned, 'constant recombination rate of 1.28 × 10−8 and a mutation rate of 2.36 × 10−8.' Additionally, it would be helpful to describe (where do they come from, for example) the demographic parameters in the Materials and Methods section and include the msprime code.
In Scenario C (Figure 1), using the American-Admixture demographic model, the study states that 'we consistently observed that ADMIXTURE and fastmixture perform similarly in accuracy, with results closest to the ground truth (Table 1 and Table S2).' However, I believe the standard error should also be taken into account. ADMIXTURE shows a standard error ten times smaller than fastmixture.
To present the results from Table 1 more visually, consider calculating the KL divergence between the predicted admixture values and the ground truth for each individual in each population and study for each evaluated method. This could help identify if any particular method exhibits more bias toward certain genetic backgrounds. For instance, the Neural ADMIXTURE paper observed that their method produces harder cluster predictions compared to ADMIXTURE, potentially impacting admixture proportions in mixed populations, as suggested by the authors of this study.
In the legend of Figure 2, please specify that it uses the downsampled version (I understand this applies to all methods, not just ADMIXTURE).
It would also be very interesting to evaluate the performance of the proposed algorithm concerning the hyperparameters it requires.
Title and abstract
Does the title clearly reflect the content of the article? Yes
Does the abstract present the main findings of the study? Yes
Introduction
Are the research questions/hypotheses/predictions clearly presented? Yes
Does the introduction build on relevant research in the field? Yes
Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? Yes
Are the methods and statistical analyses appropriate and well described? Yes
Results
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? Yes
Are the results described and interpreted correctly? Yes
Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? Yes
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? Yes