
The Bowmouth Guitarfish Doesn’t Travel Far: Genomic Evidence for Low Connectivity and Regional Differentiation in Rhina ancylostomus
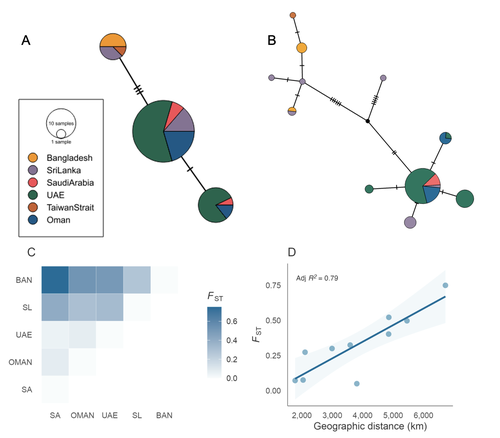
Population structure and genetic diversity of the Critically Endangered bowmouth guitarfish (Rhina ancylostomus) in the Northwest Indian Ocean
Abstract
Recommendation: posted 13 May 2025, validated 14 May 2025
Nabholz, B. (2025) The Bowmouth Guitarfish Doesn’t Travel Far: Genomic Evidence for Low Connectivity and Regional Differentiation in Rhina ancylostomus. Peer Community in Evolutionary Biology, 100792. https://doi.org/10.24072/pci.evolbiol.100792
Recommendation
Chondrichthyan fishes (sharks, rays, and chimaeras) are among the most endangered taxa on Earth. Most species exhibit life-history traits that make them particularly sensitive to both direct and indirect human impacts, especially fisheries (Jabado et al. 2024). In this context, genomic data can provide crucial insights into population structure and historical and current connectivity among populations, thereby informing conservation strategies. Increasing genomic research would also enhance our understanding of shark, ray, and chimaera diversity, including the identification of cryptic species and or subspecies.
In this study, Kipperman et al. (2025) investigate the population structure of the bowmouth guitarfish (Rhina ancylostomus), an endangered species in the understudied family Rhinobatidae. This represents the first analysis of population structure in this species. The authors combine over 3,000 nuclear SNPs, obtained using a restriction enzyme reduction-based DNA sequencing strategy, with mitochondrial data from 70 samples across six populations in the Northwest Indian Ocean. Their results reveal significant population structure driven by isolation-by-distance between eastern and western populations across most of the species' range, from Saudi Arabia to Taiwan. These findings highlight limited connectivity among populations and demonstrate the species' low dispersal capacity.
Further analyses of genetic diversity show that populations in eastern Saudi Arabia and Oman have significantly lower genetic diversity than those in the eastern part of the species’ range. The authors hypothesize that this pattern results from a westward colonization of the species from the Indo-Malayan archipelago.
The study has a few minor limitations, including unbalanced and incomplete sampling across the species’ range and a lack of detailed description of the SNP sequencing and bioinformatics pipeline, which is based on proprietary protocols. The absence of a reference genome is also a drawback. However, all these limitations are transparently acknowledged and discussed in the manuscript. Moreover, they do not undermine the robust analyses in the manuscript.
Kipperman and collaborators’ study provides important insights into the genetic structure of a critically endangered guitarfish species. Well-designed analyses such as this are essential for improving our knowledge of chondrichthyan population structures.
References
Jabado, R.W., Morata, A.Z.A., Bennett, R.H., Finucci, B., Ellis, J.R., Fowler, S.L., Grant, M.I., Barbosa Martins, A.P., & Sinclair, S.L. (eds.) (2024). The global status of sharks, rays, and chimaeras. Gland, Switzerland: IUCN https://doi.org/10.59216/ssg.gsrsrc.2024
Marja J. Kipperman, Rima W. Jabado, Alifa Bintha Haque, Daniel Fernando, P A D L Anjani, Julia LY Spaet, Emily Humble (2025) Population structure and genetic diversity of the Critically Endangered bowmouth guitarfish (Rhina ancylostomus) in the Northwest Indian Ocean. bioRxiv, ver.2 peer-reviewed and recommended by PCI Evolutionary Biology https://doi.org/10.1101/2024.03.15.585225
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
This work was generously supported by the Save our Seas Foundation and the Shark Conservation Fund. Sample collection in Oman and the UAE was supported by the United Arab Emirates University. Sample collection in Sri Lanka was supported by the Save Our Seas Foundation, the Shark Conservation Fund, the Marine Conservation and Action Fund (MCAF) of the New England Aquarium, the Ocean Park Conservation Foundation, Hong Kong [FH02_1920], and the Tokyo Cement Group.
Reviewed by anonymous reviewer 1, 06 May 2025
I am happy with the new version of the manuscripts. I recommend it for publication!
https://doi.org/10.24072/pci.evolbiol.100792.rev21Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2024.03.15.585225
Version of the preprint: 1
Author's Reply, 10 Apr 2025
Decision by Benoit Nabholz, posted 02 Jul 2024, validated 02 Jul 2024
Your manuscript was evaluated by three reviewers. Reviewer #1 and #3 are positive and all appreciated your work. I largely agree and believe that the use of a powerful genomic method to better understand the population structure of an endangered shark species is very valuable and of great importance for conservation.
Reviewer #1 offers good advice for improving your text. I noticed that you have already addressed the comment regarding the low genetic diversity of the CR region in the discussion section.
Reviewer #2 provides numerous comments and focuses on sampling issues. The limitation of not sampling the Indo-Malayan archipelago could be discussed further. For instance, you could expand on references to coastal reef shark studies if they support the reviewer's suggestion that the Indo-Malayan regions often harbor greater genetic diversity. Additionally, the imbalance in sampling between localities is noted. I suggest performing a control analysis by randomly subsampling all populations to N=4 or 5. This would allow you to test the effect of unbalanced sampling on your conclusions. Indeed, unbalanced sampling has been shown to influence structure-like analyses (Puechmaille 2016) and PCA. Finally, several comments are made regarding your discussion. While I do not fully agree with all of them, I concur that you could directly interpret your results as supporting a clinal variation from east to west with discontinuous sampling creating gaps. Therefore, I suggest modifying your text accordingly in both the results and discussion sections (e.g., lines 337-341).
Reviewer #3 is positive. He wants clarification on the potential ascertainment or allelic bias of the nuclear marker. He also adds a comment on the clinal nature of the genetic variation, which supports the modifications suggested above.
Please respond to all the comments made by the reviewers, but if you choose not to accept some of them, please justify your decision in the replies.
As a final note, I haven’t found the supplementary figures and tables to be available. Could you please make them available in the next version of your preprint?
Reference
Puechmaille SJ. 2016. The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol. Ecol. Resour. 16:608–627.
Reviewed by anonymous reviewer 1, 29 May 2024
The manuscript from Kipperman et al. investigates the population structure of the bowmouth guitarfish at both mitochondrial (control region and COI) and nuclear (DART sequencing) level. The bowmouth guitarfish represents an interesting case study due to its status (Critically Endangered in the IUCN Red list of Threatened species) and its geographic distribution.
I do not see any major flows in the data generation process and population genetics analyses. However, the sampling strategy is not optimal. I do understand that it is very difficult to perform an adequate and extensive sampling for marine species and I can imagine that adding more individuals would be impossible at this stage, but the data at hand are unfortunately limited. As consequence, conclusions do not seem highly robust and are a bit limited in scope. There are three problems for me concerning the sampling: the low number of individuals sequenced, the unbalanced sampling (67% of the whole nuclear samples comes from the same locality, UAE), the geographic locations not covering much of the total distribution and typically ignoring regions which are most likely harboring largest diversity (the Indo Malayan archipelago) following many biogeographic studies and the genetic diversity in coastal reef sharks.
I have some remarks about the discussion, which sometimes is unnecessary long and disconnected from evidence presented in this work.
Lines 320-321. It is unclear what do they mean with ‘contemporary evolutionary constraints’. Also, past directional selection should affect also the COI since the mtDNA is a single linkage block. Also it is not clear what biases in mutation type are and how should they contribute to determine the observed pattern.
Lines 328-330. First, the observed pattern may partially be driven from the unbalanced sampling. Second, it seems from their results that there is a clinal distribution of the genetic diversity, implying continuous gene flow driven by geographic distances. At best, this sentence should be reformulated by stressing the evidence of continuous variation in diversity.
Lines 330-332. I do not understand the meaning of this sentence. The analyses do not suggest the presence of two differentiated clusters but, once again, of a clinal variation. Also, Fst analyses are not extremely useful in this context given the unbalanced sampling and the very low number of individuals of some locations. I suggest removing the Fst because of this sampling scheme.
Lines 337-339. I do not understand this sentence. The authors observed linear differentiation, which is a form of population structure. I do not see the antithesis with “true population differentiation”. Please remove of reframe this sentence.
Lines 352-363. Not sure this paragraph adds much to the manuscript
Line 370. The species is present only in the IP, so clearly it would be impossible to infer its origin anywhere else. But I agree that given the sampling, it is impossible to infer its centre of origin correctly
Lines 387-388. It seems that gene flow is ongoing but this has not been formally tested (populations can be recently become isolated, for example). Not sure this sentence adds much.
Lines 390-399. This applies to any species. It is unclear why the results presented here should prompt more conservation effort than in any other species. Maintaining habitat connectivity seems to be always crucial, but this is quite unlinked from the results presented here.
Minor comment:
- Line 180: it is unclear how the bootstrap is performed. Is it the same permutation approach of Excoffier et al. 1992?
https://doi.org/10.24072/pci.evolbiol.100792.rev11Reviewed by anonymous reviewer 2, 14 May 2024
Reviewed by anonymous reviewer 3, 02 Jul 2024
The authors present a conservation genetics study of the bowmouth guitarfish, in which they use mitochondrial markers and SNPs from a nuclear SNP-typing array to assess population structure over much, but not all, of the species range.
There are not many details on the SNPtyping, for example how SNPs were ascertained, and how/whether the authors test for ascertainment or allelic bias. It would be useful to have such details to be order to assess the structure results from nuclear markers.
Sample sizes are uneven among putative demes, and often small, which the authors acknowledge and address.
The pattern of weak structuring reflects recent findings in some dolphin species (Gose et al. 2024) by a team that includes the lead author of this study, and perhaps this is more the norm in this high connectivity environment. I did wonder whether the apparent break point in the structuring between SL and UAE is due to the lack of sampling and geographic distance between the two, so that the reality is this even more of a cline than suggested, rather than a case of clear structuring.
Overall, the authors have performed a suite of analyses that complement each other, and have judiciously assessed the strengths and weaknesses of the resulting findings of each. This study is clearly of high importance for the conservation efforts to manage this critically endangered species.
I therefore recommend the study for publication.
https://doi.org/10.24072/pci.evolbiol.100792.rev13









