
Studying genetic antagonisms as drivers of genome evolution
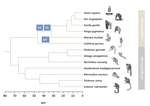
Evolutionary stasis of the pseudoautosomal boundary in strepsirrhine primates
Abstract
Recommendation: posted 22 September 2020, validated 22 September 2020
Joron, M. (2020) Studying genetic antagonisms as drivers of genome evolution. Peer Community in Evolutionary Biology, 100108. https://doi.org/10.24072/pci.evolbiol.100108
Recommendation
Sex chromosomes are special in the genome because they are often highly differentiated over much of their lengths and marked by degenerative evolution of their gene content. Understanding why sex chromosomes differentiate requires deciphering the forces driving their recombination patterns. Suppression of recombination may be subject to selection, notably because of functional effects of locking together variation at different traits, as well as longer-term consequences of the inefficient purge of deleterious mutations, both of which may contribute to patterns of differentiation [1]. As an example, male and female functions may reveal intrinsic antagonisms over the optimal genotypes at certain genes or certain combinations of interacting genes. As a result, selection may favour the recruitment of rearrangements blocking recombination and maintaining the association of sex-antagonistic allele combinations with the sex-determining locus.
The hypothesis that sexually antagonistic selection might drive recombination suppression along the sex chromosomes is not new, but there are surprisingly few studies examining this empirically [1]. Support mainly comes from the study of guppy populations Poecilia reticulata in which the level of sexual dimorphism (notably due to male ornaments, subject to sexual selection) varies among populations, and was found to correlate with the length of the non-recombining region on the sex chromosome [2]. But the link is not always that clear. For instance in the fungus Microbotryum violaceum, the mating type loci is characterized by adjacent segments with recombination suppression, despite the near absence of functional differentiation between mating types [3].
In this study, Shearn and colleagues [4] explore the patterns of recombination suppression on the sex chromosomes of primates. X and Y chromosomes are strongly differentiated, except in a small region where they recombine with each other, the pseudoautosomal region (PAR). In the clade of apes and monkeys, including humans, large rearrangements have extended the non recombining region stepwise, eroding the PAR. Could this be driven by sexually antagonistic selection in a clade showing strong sexual differentiation?
To evaluate this idea, Shearn et al. have compared the structure of recombination in apes and monkeys to their sister clade with lower levels of sexual dimorphism, the lemurs and the lorises. If sexual antagonism was important in shaping recombination suppression, and assuming lower measures of sexual dimorphism reflect lower sexual antagonism [5], then lemurs and lorises would be predicted to show a shorter non-recombining region than apes and monkeys.
Lemurs and lorises were terra incognita in terms of genomic research on the sex chromosomes, so Shearn et al. have sequenced the genomes of males and females of different species. To assess whether sequences came from a recombining or non-recombining segment, they used coverage information in males vs females to identify sequences on the X whose copy on the Y is absent or too divergent to map, indicating long-term differentiation (absence of recombination). This approach reveals that the two lineages have undergone different recombination dynamics since they split from their common ancestor: regions which have undergone further structural rearrangements extending the non-recombining region in apes and monkeys, have continued to recombine normally in lemurs and lorises. Consistent with the prediction, macroevolutionary variation in the differentiation of males and females is indeed accompanied by variation in the size of the non-recombining region on the sex chromosome.
Sex chromosomes are excellent examples of how genomes are shaped by selection. By directly exploring recombination patterns on the sex chromosome across all extant primate groups, this study comes as a nice addition to the short series of empirical studies evaluating whether sexual antagonism may drive certain aspects of genome structure. The sexual selection causing sometimes spectacular morphological or behavioural differences between sexes in many animals may be the visible tip of the iceberg of all the antagonisms that characterise male vs. female functions generally [5]. Further research should bring insight into how different flavours or intensities of antagonistic selection can contribute to shape genome variation.
References
[1] Charlesworth D (2017) Evolution of recombination rates between sex chromosomes. Philosophical Transactions of the Royal Society B: Biological Sciences, 372, 20160456. https://doi.org/10.1098/rstb.2016.0456
[2] Wright AE, Darolti I, Bloch NI, Oostra V, Sandkam B, Buechel SD, Kolm N, Breden F, Vicoso B, Mank JE (2017) Convergent recombination suppression suggests role of sexual selection in guppy sex chromosome formation. Nature Communications, 8, 14251. https://doi.org/10.1038/ncomms14251
[3] Branco S, Badouin H, Vega RCR de la, Gouzy J, Carpentier F, Aguileta G, Siguenza S, Brandenburg J-T, Coelho MA, Hood ME, Giraud T (2017) Evolutionary strata on young mating-type chromosomes despite the lack of sexual antagonism. Proceedings of the National Academy of Sciences, 114, 7067–7072. https://doi.org/10.1073/pnas.1701658114
[4] Shearn R, Wright AE, Mousset S, Régis C, Penel S, Lemaitre J-F, Douay G, Crouau-Roy B, Lecompte E, Marais GAB (2020) Evolutionary stasis of the pseudoautosomal boundary in strepsirrhine primates. bioRxiv, 445072. https://doi.org/10.1101/445072
[5] Connallon T, Clark AG (2014) Evolutionary inevitability of sexual antagonism. Proceedings of the Royal Society B: Biological Sciences, 281, 20132123. https://doi.org/10.1098/rspb.2013.2123
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Evaluation round #3
DOI or URL of the preprint: https://doi.org/10.1101/445072
Version of the preprint: 5
Author's Reply, 07 Sep 2020
We have done all the requested changes (include Table S2 in the main text + reformat the ms using the PCIEvolBiol template). Please visit the version 6 of our preprint on biorxiv: https://www.biorxiv.org/content/10.1101/445072v6 (will be available in the next few hours)
Decision by Mathieu Joron, posted 03 Sep 2020
Dear Gabriel Marais
Thanks for submitting the revision of your manuscript “Evolutionary stasis of the pseudoautosomal boundary in strepsirrhine primates”. Looking at your changes and your rebuttal letter, I am of course still willing to recommend your manuscript.
However, now that I see the (formerly missing) table S2 and its content, I find it odd that this section is separated from the main study, and with non-tabular material such as methods description and references only appearing in cells of an excel file. Reading your manuscript again, it appears that the phenotypic analysis stands as an integral part of the logic linking sexual antagonism to your comparative genomics data. So I consider that table S2 should probably be moved to the main manuscript (as a new “Table 1” for instance), rather than appearing as a supplementary.
Making this change seems pretty painless : the text appearing in S2 can be transferred at the end of the main methods section and the literature cited combined within the main list of references. By doing so, the data table, which is of small size, becomes the only tabular material in the document and can easily be pasted in the main PDF. Furthermore, this would not remove the emphasis on the genomic patterns, nor would it require any change in manuscript structure or distract the reader in any way, since the methods are appended after the discussion and with clear subheading. It would improve coherence and long-term accessibility of the data to the reader, which cannot be guaranteed if the phenotypic analysis is separated from the main text.
Apart from this I am satisfied with you changes and I really like you manuscript. I hope you will agree to those slight changes, and am looking forward to the final version.
Mathieu Joron
Evaluation round #2
DOI or URL of the preprint: 10.1101/445072
Version of the preprint: 4
Author's Reply, 27 Aug 2020
see file
Decision by Mathieu Joron, posted 19 Aug 2020
Dear Gabriel Marais
Your revised manuscript was reviewed by three referees, whose comments are pasted below. All referee consider that your manuscript addresses an interesting question and provides a plausible interpretation with solid data and analysis. I agree with them that the manuscript is much improved and that you responded adequately to the earlier comments. Overall I would be glad to recommend your manuscript but two referees and myself still have a number of suggestions and requests for clarifications which seem important to address before formal acceptance.
1- The analysis of sexual dimorphism as a proxy for sexual antagonism is done adequately. However, the methods are totally absent from the manuscript and the supplementary table S2 is also absent (an excel file apparently). Since the methods are explained in the rebuttal letter, I only became aware of this when reading the actual reviews. Of course table S2 and the corresponding methods section are needed in the revised version.
2- Both referees 2 and 3 still point to your interpretation (differences in sexually-antagonistic selection explaining differences in recombination suppression) being speculative, even though the ideas are carefully expressed and you acknowledge other processes. In my view, the logic could be more carefully explained. Indeed, looking for differences in sexual selection as a support for a role of SA selection leads the reader to understand that the differences in SA selection could be sufficient to explain the genomic signals, yet this is unclear and merits a careful explanation. SA selection is multifaceted and sexes in mammals differ in a large number of traits, a fraction of which is directly affected by sexual selection (as measured with morphological markers), so clades with little sexual selection might still undergo significant SA selection throughout the development of males vs females. Perhaps the assumptions associated with using sexual selection as a proxy for SA selection should be briefly explained in order to make the underlying logic clearer, and perhaps the conclusions easier to appreciate.
3- Reviewer 3 has listed a number of additional points, mainly about clarity and logic, and those should be addressed in your revision.
I think the above suggestions should be easy to address by relatively light changes to the text. I am looking forward to reading your final version.
Mathieu Joron
Additional requirements of the managing board:
As indicated in the 'How does it work?’ section and in the code of conduct, please make sure that:
-Data are available to readers, either in the text or through an open data repository such as Zenodo (free), Dryad or some other institutional repository. Data must be reusable, thus metadata or accompanying text must carefully describe the data.
-Details on quantitative analyses (e.g., data treatment and statistical scripts in R, bioinformatic pipeline scripts, etc.) and details concerning simulations (scripts, codes) are available to readers in the text, as appendices, or through an open data repository, such as Zenodo, Dryad or some other institutional repository. The scripts or codes must be carefully described so that they can be reused.
-Details on experimental procedures are available to readers in the text or as appendices.
-Authors have no financial conflict of interest relating to the article. The article must contain a "Conflict of interest disclosure" paragraph before the reference section containing this sentence: "The authors of this preprint declare that they have no financial conflict of interest with the content of this article." If appropriate, this disclosure may be completed by a sentence indicating that some of the authors are PCI recommenders: “XXX is one of the PCI XXX recommenders.”
Reviewed by Qi Zhou, 22 Jul 2020
I appreciate the authors spent the time to collect sexual dimorphism data and performed the correlation analyses with and without considering the phylogeny. I think their answers addressed my previous questions and the ms should be accepted for publication.
https://doi.org/10.24072/pci.evolbiol.100184.rev21Reviewed by anonymous reviewer 3, 02 Jul 2020
This manuscript examines sex chromosome evolution in strepsirrhine primates, specifically looking for evidence of changes in the extent of the pseudoautosomal region. They find evidence that this boundary has been very stable over time, suggesting little decrease in the extent of recombination between the X and Y over time, especially compared to other primates. They interpret this finding as being consistent with low levels of sexual antagonism within strepsirrhine primates, and support this interpretation with an analysis showing that the degree of sexual dimorphism is lower in strepsirrhines than in other primates.
I was not one of the original reviewers of this manuscript, but after reading their comments and the authors' response to them, it seems like the revised version has successfully addressed the criticisms that were raised in the first round of review. I did feel that the last sentence of the abstract is a bit strongly worded ("Our work supports the view that sexually antagonistic mutations have influenced the evolution of sex chromosomes in primates"), but the main text provides a more nuanced view and acknowledges that other explanations are possible.
Unfortunately I was not able to find any information about the analysis of sexual dimorphism and testes size anywhere other than the response to reviewers. Table S2 with the results of the analysis was missing from the supplementary information files, and there was no detailed description of how the analysis was carried out. This is presumably an oversight on the part of the authors, but it's a shame since this seems like it was a nice addition to the manuscript, and I currently can't evaluate the robustness of these results.
https://doi.org/10.24072/pci.evolbiol.100184.rev22Reviewed by anonymous reviewer 2, 02 Jul 2020
This is an interesting analysis of the evolution of the pseudo-autosomal region in primates, which demonstrates that the suppression of recombination between the X and the Y chromosomes has proceeded along different trajectories in the two main lineages of primates. In haplorrhines, there has been further suppression of recombination leading to the formation of two additional strata, while in strepsirrhines, there has been no further suppression of recombination since these lineages diverged. The patterns of recombination suppression are consistent with the presence of high levels of sexual dimorphism in haplorrhines and low levels of sexual dimorphism in strepsirrhines. Using levels of sexual dimorphism as a proxy for the intensity of sexually antagonistic selection, the authors conclude that selection for linkage between the sex-determination locus and alleles with sexually antagonistic effects might have driven suppression of recombination and the formation of the two additional strata in haplorrhines. I find this an intriguing hypothesis that lays the groundwork for additional studies. Although these conclusions remain speculative, the authors are careful in their conclusions and clear about the limitations. They have responded well to the previous reviews.
Nonetheless, the manuscript could be more clearly written in places, and so I provide some suggestions here for revisions to improve the clarity of the presentation.
- P4, L25 - P5, L1: change “a ratio of 0.5 the X-specific region” to “a ratio of 0.5 should indicate the X-specific region”
- P5, L15 and P6, L2: perhaps change “human strata 4, 5 and PAR1” to “PAR1 and human strata 4 and 5”
- P5, L17: change “and agreeing with” to “in agreement with”
- P5, L23 - P6, L1: the sentences here seem out of order. Perhaps start by saying why the fragmented assembly cannot identify the size of the PAR or autosomal to PAR translocations, and then state that an improved, de novo assembly based on long-read sequencing would allow you to confirm one of these alternatives.
- P6, L3: “the location of the PAB is conserved” – conserved to what? The PAB is different in grey mouse lemur and humans. Either delete this phrase or be more specific.
- P6, L5: change “only available one” to “only one available”
- P6, L12: change “PAR1 and strata 4 and 5 in humans”
- P6, L13: Interestingly, in Figure S2, the location of the PAB seems to differ between lorises and lemurs, but I assume this is simply due to differences in the X assembly?
- P6, L14: change “no recombination suppressing event” to “no suppression of recombination between the X and the Y”
- P6, L16 - P7, L15: here, I suggest changing the order of presentation; currently it is very confused. First, I would present the density of SNPs to show that you did not miss any strata. Simply move this paragraph up. Then, I would have a separate paragraph describing the rates of strata formation, with the conclusion that the rate is higher in haplorrhines. Finally, you can end with a paragraph describing the analyses of sexual dimorphism and say that the higher levels of sexual dimorphism in haplorrhines is consistent with sexual antagonism driving higher rates of strata formation in this group. For me, this is a nice logical presentation that builds the results to a clear statement of your major conclusions at the end.
- Discussion, P7, L21: delete “and this is validated by a statistical test”. Also, in this paragraph, the biggest caveat of all these analyses is that it is a comparative study, and correlation does not equal causation! Even if you had a large phylogenetic study, this remains a caveat.
- P8, L6-9: the evidence for expansion of loss of recombination on the guppy Y chromosome in high predation populations is not supported by the work of the Charlesworth group, and I think this needs to be acknowledged here (e.g. Bergero et al. 2019 PNAS).
- P8, L13: please define “a process of attrition” and change “DNA repeats accumulation” to “accumulation of DNA repeats”.
- P8, L14-15: this is a challenging sentence to read. Perhaps change to “In Microbotryum violaceum, strata are found on the mating type chromosomes despite the fact that this species only has mating types and not sexes, such that sexual antagonism is absent”.
- P8, L22-23: What is the evidence that there is more suppression of recombination in red-bellied lemur than in the other species? I do not see this in Figure 1 or the supplementary figures?
- P9, L24-25: Perhaps cite a nice recent paper in Science that identifies the genetic basis of sexual dichromatism in canaries (Gazda et al Science 12 June 2020)
- In Figure 1 (as well as supplementary figures), can you add the common names of the species (used in the manuscript) to the scientific names (used in the figures) so that the reader can easily find the appropriate panel without needing to look at Table S1.
- P12, L5-6: How can a major allele frequency be 0.3x the site coverage? For example, if the site coverage is 10, then the major allele frequency would be 3. This does not make sense. Also, these analyses do not seem very robust, as the older strata are also not identified these analyses. Most of the SNP densities across the X chromosome seem to fall within the autosomal range?
Evaluation round #1
DOI or URL of the preprint: 10.1101/445072
Version of the preprint: 3
Author's Reply, 28 May 2020
see file.
Decision by Mathieu Joron, posted 01 Apr 2019
Dear Rylan Shearn and Gabriel Marais
Your manuscript was reviewed by two referees whose comments are attached. Both referees found your study very interesting, remarking that it contributes nicely to sex chromosome research and should be of broad interest to evolutionary biologists. The role of selection in recombination suppression is a timely subject and this study brings new and interesting data using sex chromosome strata formation, and contrasting the known structure of sex chromosomes in haplorrhine primates to their understudied sister group, the Strepsirrhines.
However, based on those reviews and my own reading, I think that your manuscript requires a few improvements to lead to a recommendation. Both reviewers find the link between the lack of strata 4 and 5 in Strepsirrhines and the lower level of sexually antagonistic selection acting in this group still tenuous, and would have expected a quantitative evaluation of sexual dimorphism, or a discussion of alternative mechanisms. After all, the two groups may vary in many ways which could affect strata formation. Therefore, it seems essential to improve the argument for or against isolating SA selection as the factor causing differences in chromosome structure. Referee 1 would like more details on the use of male/female SNP density of 0.5 to identify regions with suppressed recombination, and the paper would benefit from a critical discussion of the factors affecting this metric and determining the expected values under recombination suppression. Finally, following Referee 2, providing further details on how the PAB and the segments corresponding to strata 4 and 5 in Humans vary among Strepsirrhines would also improve the manuscript.
I would therefore invite you to submit a revised version taking those suggestions into account, and explaining in details how you have dealt with the points raised by the reviewers.
Thank you for sending this interesting work to PCI Evol Biol. I look forward to seeing the revision.
Best regards,
Reviewed by anonymous reviewer 1, 25 Feb 2019
Shearn et al conduct a comparative analysis of sex chromosome evolution across two primate groups to test the relationship between sexual dimorphism, a proxy for sexual conflict, and recombination suppression. This is a hot topic at the moment. They find that there is a higher rate of strata formation in haplorrhine primates than strepsirrhines, a direction consistent with the role of sexual conflict in halting recombination. I enjoyed reading the manuscript and think it makes a valuable contribution to sex chromosome research. My comments are below:
Can the authors comment about when sexual dimorphism evolved in haplorrhines? This essential to interpret the results as the argument that sexual conflict promoted the evolution of S4 and S5 only holds if the common ancestor of haplorrhines was sexually dimorphic.
The discussion of alternative processes promoting recombination suppression is a bit light. Eg. Úbeda F, Patten MM, Wild G (2015) On the origin of sex chromosomes from meiotic drive. Proc Biol Sci 282:20141932 Matsumoto T, Yoshida K, Kitano J (2017) Contribution of gene flow to the evolution of recombination suppression in sex chromosomes. J Theor Biol 431:25–31
I don’t quite follow the use of SNP density to identify recent strata. Why is the sum of SNPs divided by four? Do you really expect 0.5 for the old strata? I am bit perplexed by the results in the SI. I agree that in regions where the Y has largely degenerated, we might expect SNP density to be lower in males as the X is effectively hemizygous in males. But this will depend on the level of standing genetic variation in the population, and whether females are homozygous for the X. I don’t understand how all of the X has a SNP density ratio of 0.5? I would expect the ratio to be much closer to 1.
For the methods further detail is necessary on: a. Whether any trimming steps on the reads was taken b. What thresholds were imposed on SNP calling ie minor allele frequency
The colour scheme of Fig 1 need to be improved for clarity. a. Firstly, mPAR needs to be defined in the figure legend, I assume mPAR refers to the ancestral PAR (what does the m mean)? At the moment it is red, which is the same colour as s5. Therefore, it looks like s5 is at the beginning of the X for the 7 species studied when I believe the authors are trying to show this is ancestral PAR. b. Similarly, misA needs to be defined, I assume it means misassembled autosomal scaffolds? But currently it is grey which makes these regions look like PAR.
Abstract L13 ‘Moved’ seems a bit unspecific in this sentence, I would replace with extended PAR. P3 L12 I’m not aware of any direct evidence that recombination between the primate X and Y stopped instantaneously as ‘at once’ suggests. I would remove this. P3 L14 I am not sure what process is referred to in this sentence. P7 L12 I believe Wright et al showed extension of recombination suppression not additional strata P7 L17 What does ‘process of erosion’ refer to? P8 L24 Typo – should be in not on P9 L6 Typo – should be cell P9 L10 Typo – should be samples P9 L14 Typo – should be on not onto P10 L4 Should be gap after bracket P10 L6 Remove ; P10 L15 Remove ) Fig 1 Legend Missing of after Identification Fig 1 Legend What does the sentence starting ‘Strata in humans using definition of strata shown …’ mean? Why does coverage ratio approach 1.5 in some species?
https://doi.org/10.24072/pci.evolbiol.100184.rev11Reviewed by Qi Zhou, 25 Feb 2019
Sex chromosomes show a dynamic composition even between related species regarding their regions with or without recombination. The sex-linked regions with recombination is called pseudoautosomal regions (PAR), and this work by Rylan et al. found that strepsirrhines (lemurs and lorises) share a longer PAR than other primates (apes and monkeys), and they explained this pattern by suggesting a lower level of sexual antagonistic selection in the former than the latter. It is a great effort to characterize the PAR of all seven species, and the topic should be of broad interest to evolutionary biologists. However, I hope the authors provide more quantitative measurements of the level of sexual antagonistic (SA) selection ongoing in the strepsirrhines. Because after all, the experimental evidence for SA selection causing the recombination loss is quite rare. Guppy is an excellent example because the previous comparison by Alison et al was performed between two closely related populations with a very clear indication of sexual selection (the color), thus the association between the expanded recombination suppression region vs. different color (different degree of SA selection) can be interpreted as the suggestive evidence. The author talked briefly in the introduction about sexual size dimorphism, and how exactly was the degree of strepsirrhines compared to that of haplorrhines? How about the mating-system in these two groups of haplorrhines? I only know the mating system is associated with the relative testis size of primates, which is indicative of the strength of sexual selection. Second, I am not sure if the author has identified a scaffold that shows a switch between the M:F ratio of 1 vs. 0.5. The authors mentioned scaffolds that show a M:F ratio of 1 correspond to strata 4 & 5 in human, this only indicate there were no recombination loss after strata 3 formed in strepsirrhines. A process called attrition, which as mentioned by the authors is the gradual erosion of PAR by repetitive elements has been reported in cow, and in fact, many mammalian PABs are characterized by insertion of TEs (reviewed by Terje et al. 2015). Has the author found a similar pattern? Or are the PABs of the seven species actually aligned to each other? Finally, just curious, has the author managed to assemble any Y-linked genes, and are they any different from those that have been assembled for other mammals?
Some other minor comments:
introduction: line 1, 'strongly' maybe changed to 'very';
line 9, the master male-determining gene in Eutherian mammals, as platypus does not share the same sex chromosome and master sex determining gene;
line 15, I suggest to emphasize here that all eutherian mammals share strata 1-3;
page 4, line 21, the same copy number of autosomes;
page 5, line 16, are there any read pairs that span the X-specific region and the autosome like region? If so, there should help to clarify if there are potential recent fusion or just assembly errors;
page 6, I am not sure about the N50 of gray mouse lemur, but there is also a blue-eyed black lemur genome published with about 400kb N50 size, would it be better to use that as reference?










