
Diverse outcomes in cheese fungi domestication
 based on reviews by Delphine Sicard and 1 anonymous reviewer
based on reviews by Delphine Sicard and 1 anonymous reviewer
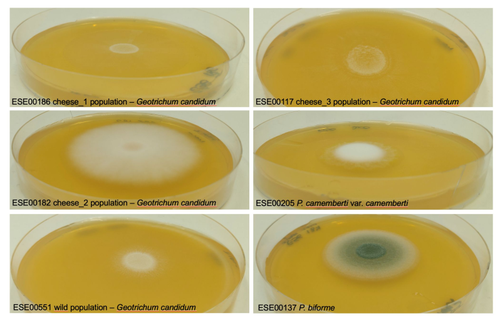
Domestication of different varieties in the cheese-making fungus Geotrichum candidum
Abstract
Recommendation: posted 23 March 2023, validated 24 March 2023
Fraïsse, C. (2023) Diverse outcomes in cheese fungi domestication. Peer Community in Evolutionary Biology, 100592. https://doi.org/10.24072/pci.evolbiol.100592
Recommendation
Domestication is a complex process that imprints the demography and the genomes of domesticated populations, enforcing strong selective pressures on traits favourable to humans, e.g. for food production [1]. Domestication has been quite intensely studied in plants and animals, but less so in micro-organisms such as fungi, despite their assets (e.g. their small genomes and tractability in the lab). This elegant study by Bennetot and collaborators [2] on the cheese-making fungus Geotrichum candidum adds to the mounting body of studies in the genomics of fungi, proving they are excellent models in evolutionary biology for studying adaptation and drift in eukaryotes [3].
Bennetot et al. newly showed with whole genome sequences that all G. candidum strains isolated from cheese form a monophyletic clade subdivided into three genetically differentiated populations with several admixed strains, while the wild strains sampled from diverse geographic locations form a sister clade. This suggests the wild progenitor was not sampled in the present study and calls for future exciting work on the domestication history of the G. candidum fungus. The authors scanned the genomes for footprints of adaptation to the cheese environment and identified promising candidates, such as a gene involved in iron uptake (this element is limiting in cheese). Their functional genome analysis also provides evidence for higher contents of transposable elements in cheese-making strains, likely due to relaxed selection during the domestication process.
This paper is particularly impressive in that the authors complemented the population genomic approach with the phenotypic characterization of the strains and tested their ability to outcompete common fungal food spoilers. The authors convincingly showed that cheese-making strains display phenotypic differences relative to wild relatives for multiple traits such as slower growth, lower proteolysis activity and a greater amount of volatiles attractive to consumers, these phenotypes being beneficial for cheese making.
Finally, this work is particularly inspiring because it thoroughly discusses convergent evolution during domestication in different cheese-associated fungi. Indeed, studying populations experiencing similar environmental pressures is fundamental to understanding whether evolution is repeatable [4]. For instance, all three cheese populations of G. candidum exhibit a lower genetic diversity than wild populations. However, only one population displays a stronger domestication syndrome, resembling the Penicillium camemberti situation [5]. Furthermore, different cheese-making practices may have led to varying situations with clonal lineages in non-Roquefort P. roqueforti and P. camemberti [5, 6], while the cheese-making G. candidum populations still harbour some diversity. In a nutshell, Bennetot's study makes an important contribution to evolutionary biology and highlights the value of diversifying our model organisms toward under-represented clades.
REFERENCES
[1] Diamond J (2002) Evolution, consequences and future of plant and animal domestication. Nature 418: 700–707. https://doi.org/10.1038/nature01019
[2] Bennetot B, Vernadet J-P, Perkins V, Hautefeuille S, Rodríguez de la Vega RC, O’Donnell S, Snirc A, Grondin C, Lessard M-H, Peron A-C, Labrie S, Landaud S, Giraud T, Ropars J (2023) Domestication of different varieties in the cheese-making fungus Geotrichum candidum. bioRxiv, 2022.05.17.492043, ver. 4 peer-reviewed and recommended by Peer Community in Evolutionary Biology. https://doi.org/10.1101/2022.05.17.492043
[3] Gladieux P, Ropars J, Badouin H, Branca A, Aguileta G, de Vienne DM, Rodríguez de la Vega RC, Branco S, Giraud T (2014) Fungal evolutionary genomics provides insight into the mechanisms of adaptive divergence in eukaryotes. Mol. Ecol. 23: 753–773. https://doi.org/10.1111/mec.12631
[4] Bolnick DI, Barrett RD, Oke KB, Rennison DJ, Stuart YE (2018) (Non)Parallel evolution. Ann. Rev. Ecol. Evol. Syst. 49: 303–330. https://doi.org/10.1146/annurev-ecolsys-110617-062240
[5] Ropars J, Didiot E, Rodríguez de la Vega RC, Bennetot B, Coton M, Poirier E, Coton E, Snirc A, Le Prieur S, Giraud T (2020) Domestication of the Emblematic White Cheese-Making Fungus Penicillium camemberti and Its Diversification into Two Varieties. Current Biol. 30: 4441–4453.e4. https://doi.org/10.1016/j.cub.2020.08.082
[6] Dumas, E, Feurtey, A, Rodríguez de la Vega, RC, Le Prieur S, Snirc A, Coton M, Thierry A, Coton E, Le Piver M, Roueyre D, Ropars J, Branca A, Giraud T (2020) Independent domestication events in the blue-cheese fungus Penicillium roqueforti. Mol Ecol. 29: 2639–2660. https://doi.org/10.1111/mec.15359
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
This work was funded by the Artifice ANR-19-CE20-0006-01 ANR grant to J.R. In addition, a NSERC-Discovery Grant supported the scholarship of V.P and data generation, and is held by S. Labrie (RGPIN-2017-06388)
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2022.05.17.492043
Version of the preprint: 3
Author's Reply, 20 Feb 2023
Decision by Christelle Fraïsse , posted 21 Oct 2022, validated 26 Oct 2022
, posted 21 Oct 2022, validated 26 Oct 2022
Thank you for your patience. Two reviewers have provided constructive and thorough comments on your preprint. Both agree that the study is of high quality, and after reading the manuscript, I agree with their statement. I especially appreciated that domestication was tackled from a phenotypic and genomic perspective – this represents an impressive amount of work. I am convinced that this work will interest the evolutionary biology community.
The reviewers made a number of helpful, often concordant, suggestions for improvements that must be addressed in a revision. In particular:
1) They criticize the interpretation of the data for the Cheese_2 population as revealing a more advanced stage of domestication. Evidence for human-mediated selection to be more substantial in that cheese population than others should be demonstrated more clearly. Moreover, Reviewer 1 suggests considering other forces than selection in the evolution of the cheese populations, including migration and genetic drift.
2) Another issue raised by the two reviewers is the under-sampling of the wild strains. This bias could have consequences for interpreting the data (in particular, if the wild strains were sampled in a single geographic area), so it should be handled with caution. Reviewer 2 noted that an outcome that could follow is the misinterpretation of the cheese clade as being derived from the wild clade (see their specific comments on that point). Determining if cheese-making practices (Reviewer 1) could explain the differentiation between the three cheese clades may help understand whether this genetic structure within cheese strains has pre-existed domestication.
3) Both reviewers note the difficulty of interpreting FST when measuring the extent of population divergence. Moreover, Reviewer 2 questions the method used to detect candidate regions under adaptation. Instead of considering the 5% most extreme values of Dxy and Pi, which is expected under neutrality, you could only consider those regions that pop up as extremes in both Dxy and Pi scans as candidates. Otherwise, I recommend running a method that scans for selective sweeps.
4) I agree with Reviewer 1 that the comparison with Penicillium should be better justified and moved into a single paragraph in the Discussion section. A more in-depth discussion of the convergence process would help clarify the importance of comparing with Penicillium.
5) Reviewer 2 suggests running a demographic analysis to test for an absence of a substantial bottleneck during the domestication of G. candidum. I think there is no need to run such a type of analysis; however, Tajima’s D could be calculated to clarify this claim.
I am looking forward to receiving your revised preprint.
With best regards,
Christelle Fraïsse.
Reviewed by Delphine Sicard, 19 Oct 2022
This manuscript reported the analysis of Geotrichum candidum domestication in cheese making. This MS expands the studies on the domestication of fungi in fermented products. It presents a set of results on the genomic diversity of the fungus and its phenotypic diversity. It shows that three groups of cheese populations have diverged genetically and phenotypically from populations with wild and mixed-origin and revealed some interesting features on the evolutionary trajectories of the cheese populations. I found that the work done was adequate with the scope of the manuscript. The analysis of the interactions between Geotrichum candidum and its competitors is very original in a study on domestication. My comments are just made to improve the manuscript.
Majors :
I suggest to use the term domestication with more care. Please define domestication and discuss the results with respect to selection by cheese making practices and to migration via the use of industrial strains. Similarly, the paper refers several times to phenotypic convergence but the processes underlying this convergence should be explained. In other worlds, it would be interesting to propose the different scenarios that could explain these convergences.
We are left with not knowing if the Geotrichum candidum cheese populations have adapted to the high lipid content of cheese. The lack of lipolysis signature of domestication raised questions. The maladaptation of cheese populations to cheese agar medium, and to salt is intriguing and would deserve additional comments. Other hypothesis than evolutionary constraints could be suggested.
More generally, the involvement of evolutionary forces other than selection in the evolution of the cheese populations should be further discussed. I would expect migration, especially those associated with the sale of starter strains. I would also expect drift in environments that are often disinfected.
In terms of structure, there are several elements of discussion in the results, especially the comparison with Penicillium. I would move them into the discussion in order to reduce and clarify results and enrich the discussion.
Title – I would not call a group of genetically closely related fungal strains a variety. To me, a variety refers to a group of genotypes (most often a single genotype) that has been consciously selected by human.
Minor
L28-29: “The genetic diversity …was high”: high compared to what ?
L29:. I would not state in such determine way that the data indicates a lack of strong bottleneck because of the sampling biases. There are many more cheese strains than strains coming from elsewhere.
L32 “attractive” for who ?
L34 what do you mean by “a more advanced state of domestication” ? I don’t think the data allows the quantification of the response to selection . Moreover, the decrease in genetic diversity is not a signature of domestication. Several studies have now shown that domestication may also lead on the contrary to a diversification, at least for some traits or genetic clusters.
L74-75: S. cerevisiae has been domesticated to make a large number of fermented products beyond beer, wine and bread. Please complete and cite either a complete set of recent reviews on Saccharomyces cerevisiae domestication or a complete set of major research papers in the field. I would go for research papers and add at least Barbosa et al. 2018 (sake), Bigey et al. current biology, 2021 (bread), Gonçalves et al. current biology, 2016 (beer, wine), Ludlow et al. current biology, 2016 (coffe, cacao)
L105-if you really want to use it, define degeneration and cite a reference. Personally, I find it more informative to speak about accumulation of deleterious mutation if this is what is meant.
L125….--it is a repeat of the abstract. If you wish to add a summary of the results at the end of the introduction, it should be more concise and differ from the abstract.
L186- clarify “cheese type distribution”
L189-103- move the comparison with Peniccilium to the discussion. In addition, FST are used to measure the extent of divergence among populations of the same species relative to the net genetic diversity within the species. To compare species, absolute measures of divergence between populations should be used in preference to relative measures such as FST. (Charlesworth’s paper and others, DOI: 10.1093/oxfordjournals.molbev.a025953)
L232-233: avoid suggestion in the results, move to the discussion
L243: I can’t see the yellow Figure 1B.a., and therefore the position of commercial strains on the tree. Please, complete the Figure 1 legend as well
L247- 1,200 SNPs: how did you choose this threshold ?
L252: move to the discussion
L242-254 or elsewhere: I was not able to find any information on ploidy. Are all the strains haploid ? please, add the information.
L350-avoid example on Penicillium in the results, move to the discussion
L353-356- what does “harsh conditions” mean ? this part should go to the discussion. Furthermore, the relaxed selection hypothesis applies to industrial strains but does it really apply to non-industrial cheese strains?
L357-I would have expected “local adaptation”, i.e. a higher growth on cheese media of cheese strains compared to wild strains. I would discuss this result further in the discussion
L369- move the convergence analysis to the discussion session.
L411: delete “known to be key compounds in fermented beverages such as wine and beer”
L452-456: discussion
L485 / L498: Why do you compare the Geotrichum candidum cheese populations diversity with the ones of Penicillium sp. ? did you have any prediction based on the use of these species for cheese making ? What does the Peniccilium and Geotrichum diversity comparison brings ? I would made a single paragraph comparing both genera rather than speak about it all over the discussion. This would allow to better show the hypothesis you have to explain their evolutionary trajectories.
L503-506: the secondary domestication hypothesis can be discussed in a more general context of domestication. There is the example of the cachaça S. cerevisiae populations (Barbosa et al 2018) but other examples could also be added.
Does cheese making practices could explain the differentiation in three groups of the cheese strains ?
L533-L534: What do you mean by “ first step of domestication” ? wby “a more advanced state of domestication” ? There is no dating here, no analysis of evolutionary dynamics. I think you can’t tell where the cheese populations are on the adaptive peak. You may also have several adaptive peaks.
L555- Why is “convergence” an important question in evolution ? a deeper discussion on the interest of studying convergence would be interesting. Explaining the process behind phenotypic convergence (standing genetic variation, de novo mutation, migration/gene flow, horizontal gene transfert) would help to understand your approach.
Figures legend-please describe in more details your figures in their legend, so they can be understood as themself.
Download the review https://doi.org/10.24072/pci.evolbiol.100592.rev11Reviewed by anonymous reviewer 1, 17 Oct 2022
Please find attached my comments.
Best regards,
Download the review https://doi.org/10.24072/pci.evolbiol.100592.rev12










