
Reconciling molecular evolution and evolutionary ecology studies: a phylogenetic reconciliation method for gene-symbiont-host systems
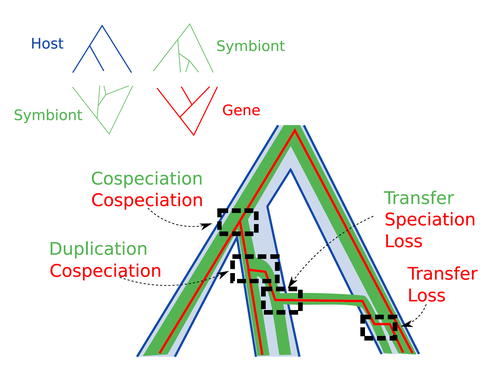
Host-symbiont-gene phylogenetic reconciliation
Abstract
Recommendation: posted 02 May 2023, validated 02 May 2023
Jousselin, E. (2023) Reconciling molecular evolution and evolutionary ecology studies: a phylogenetic reconciliation method for gene-symbiont-host systems. Peer Community in Evolutionary Biology, 100593. https://doi.org/10.24072/pci.evolbiol.100593
Recommendation
Interactions between species are a driving force in evolution. Many organisms host symbiotic partners that live all or part of their life in or on their host. Whether they are mutualistic or parasitic, these symbiotic associations impose strong selective pressures on both partners and affect their evolutionary trajectories. In fine, they can have a significant impact on the diversification patterns of both host and symbiont lineages, with symbiotic lineages sometimes speciating simultaneously with their hosts and/or switching from one host species to another. Long-term associations between species can also result in gene transfers between the involved organisms. Those lateral gene transfers are a source of ecological innovation but can obscure the phylogenetic signals and render the process of phylogenetic reconstructions complex (Lerat et al. 2003).
Methods known as reconciliations explore similarities and differences between phylogenetic trees. They have been widely used to both compare the diversification patterns of hosts and symbionts and identify lateral gene transfers between species. Though the reconciliation approaches used in host/ symbiont and species/ gene phylogenetic studies are identical, they are always applied separately to solve either molecular evolution questions or investigate the evolution of ecological interactions. However, the two questions are often intimately linked and the current interest in multi-level systems (e.g. the holobiont concept) calls for a unique model that will take into account three-level nested organization (gene/symbiont/ host) where both symbiont and genes can transfer among hosts.
Here Menet and collaborators (2023) provide such a model to produce three-level reconciliations. In order to do so, they extend the two-level reconciliation model implemented in “ALE” software (Szöllősi et al. 2013), one of the most used and proven reconciliation methods. Briefly, given a symbiont gene tree, a symbiont tree and a host tree, as in previous reconciliation models, the symbiont tree is mapped onto the host tree by mixing three types of events: Duplication, Transfer or Loss (DTL), with a possibility of the symbiont evolving temporarily outside the host phylogeny (in a “ghost” host lineage). The gene tree evolves similarly inside the symbiont tree, but horizontal transfers are constrained to symbionts co-occurring within the same host. Joint reconciliation scenarios are reconstructed and DTL event rates and likelihoods are estimated according to the model. As a nice addition, the authors propose a method to infer the symbiont phylogeny through amalgamation from gene trees and a host tree.
The authors then explore the diverse possibilities offered by this method by testing it on both simulated datasets and biological datasets in order to check whether considering three nested levels is worthwhile. They convincingly show that three-level reconciliation has a better capacity to retrieve the symbiont donors and receivers of horizontal gene transfers, probably because transfers are constrained by additional elements relevant to the biological systems. Using, aphids, their obligate endosymbionts, and the symbiont genes involved in their nutritional functions, they identify horizontal gene transfers between aphid symbionts that are missed by two-level reconciliations but detected by expertise (Manzano-Marín et al. 2020). The other dataset presented here is on the human pathogen Helicobacter pylori, which history is supposed to reflect human migration. They use more than 1000 H. pylori gene families, and four populations, and use likelihood computations to compare different hypotheses on the diversification of the host.
In summary, this study is a proof-of-concept of a 3-level reconciliation, where the authors manage to convey the applicability of their framework to many biological systems. Reported complexities, confirmed by reported running times, show that the method is computationally efficient. Without a doubt, the tool presented here will be very useful to evolutionary biologists who want to investigate multi-scale cophylogenies and it will move forward the study of associations between host and symbionts when symbiont genomic data are available.
REFERENCES
Lerat, E., Daubin, V., & Moran, N. A. (2003). From gene trees to organismal phylogeny in prokaryotes: the case of the γ-Proteobacteria. PLoS biology, 1(1), e19.
https://doi.org/10.1371/journal.pbio.0000019
Manzano-Marın, A., Coeur d'acier, A., Clamens, A. L., Orvain, C., Cruaud, C., Barbe, V., & Jousselin, E. (2020). Serial horizontal transfer of vitamin-biosynthetic genes enables the establishment of new nutritional symbionts in aphids' di-symbiotic systems. The ISME Journal, 14(1), 259-273.
https://doi.org/10.1038/s41396-019-0533-6
Menet H, Trung AN, Daubin V, Tannier E (2023) Host-symbiont-gene phylogenetic reconciliation. bioRxiv, 2022.07.01.498457, ver. 2 peer-reviewed and recommended by Peer Community in Evolutionary Biology. https://doi.org/10.1101/2022.07.01.498457
Szöllősi, G. J., Rosikiewicz, W., Boussau, B., Tannier, E., & Daubin, V. (2013). Efficient exploration of the space of reconciled gene trees. Systematic biology, 62(6), 901-912.
https://doi.org/10.1093/sysbio/syt054
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
This work was supported by the French National Research Agency (Grant ANR-19- CE45-0010 Evoluthon)
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2022.07.01.498457
Version of the preprint: 1
Author's Reply, 14 Apr 2023
Decision by Emmanuelle Jousselin, posted 08 Nov 2022, validated 09 Nov 2022
Dear authors,
Your preprint has now been reviewed by two experts, and I have also reviewed it myself. You will see that both reviewers are very positive about your manuscript and suggest that implementing a model of evolution of three nested levels (i.e. accounting for the coevolution of hosts, symbionts and their genes) represents a very useful contribution to the field of reconciliation).
They also acknowledged that the model is well implemented, its presentation is thorough and the paper clearly presented.
Being myself a user of reconciliation methods (and not a developer), I found the paper very well written and found the figures very informative and able to deliver the principle of the methods (though. I also think that such methods are needed to address reconciliation in three-way associations.
The two reviewers have nevertheless several suggestions to improve the study (see reviews). Important ones (which might require some significant inputs) concern:
-a clarification /confusion throughout the text between most likely reconciliation and maximum likelihood;
-a test of the method using data simulated under the model developed;
-some estimation (or at least discussion) of how often the model could lead to time inconsistent scenarios.
I have a few minor comments in addition to the reviewers comments and suggestions:
-concerning the use of the term coevolution: in evolutionary ecology coevolution refers to “reciprocal adaptation in interacting species”, please, if you can, use the terms cophylogeny, codivergence, cospeciation rather than coevolution that refers explicitly to the adaptive process of species.
-p2 line 50, I am not sure that reconciliation methods (using DTL model) have been implemented in Biogeographic analyses: when historical biogeography was first developed with approaches such as Brooks parsimony analyses, the method was applied to both the history of species interactions and biogeography. But biogeographic reconstructions such as the DEC approach now widely used (ref 45) are not similar to reconciliation: there is not “input tree” for geographic areas and the method is more a sophisticated method of ancestral trait reconstruction than a reconciliation. Ref 28 is a review on reconciliation methods and a comment on how they should take into account the biogeography of species rather than an example of how reconciliation methods are applied to reconstructing the biogeographic histories of species.
-in the repository : please make sure you make the output of the analyses on test datasets available (can you produce the reconciliation on Cinara aphids with Third-Kind in a jpeg or pdf format in sup mat)
I hope you find these comments helpful and look I forward to handling your revised preprint.
Please address all reviewers comments in your response.
Kind regards,
Emmanuelle Jousselin










