
LE ROUZIC Arnaud
- Évolution, Génomes, Comportement, Écologie; UMR CNRS - IRD - Université Paris-Saclay, CNRS, Gif-sur-Yvette, France
- Bioinformatics & Computational Biology, Evolutionary Theory, Genotype-Phenotype, Quantitative Genetics
Recommendations: 0
Review: 1
Areas of expertise
Genome evolution
Transposable elements
Population genetics
Quantitative genetics
Models
Individual-based simulations
Review: 1
18 Mar 2025

A gene-regulatory network model for density-dependent and sex-biased dispersal evolution during range expansions
Evolved gene-regulatory networks underlying dispersal plasticity can accelerate range expansions
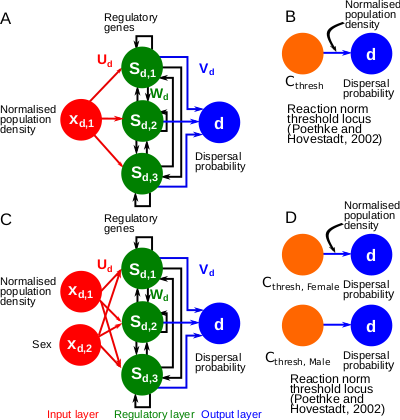
Recommended by Charles Mullon based on reviews by Arnaud Le Rouzic and 2 anonymous reviewersNatural populations rarely experience a uniform environment. Local density, resource availability, and mating opportunities often vary considerably across a population range. Theory has shown that such heterogeneity favours the evolution of density-dependent dispersal in the form of dispersal reaction norms. These models typically assume a simple genetic basis, with dispersal reaction norms encoded by a single Mendelian locus. Yet, dispersal plasticity is presumably controlled by more complex genetic architectures. Early work by Ezoe and Iwasa (1997) illustrated how evolved neural networks could generate dispersal reaction norms very similar to those predicted by Mendelian genetics.
In their manuscript, Deshpande and Fronhofer (2023) build on this work by examining how different genetic architectures influence the evolution of dispersal plasticity and its ecological consequences. They compare dispersal plasticity encoded either by a classical reaction norm controlled by a single Mendelian locus or by a gene-regulatory network following the Wagner model, where several interacting regulatory genes respond to local density cues and the individual's sex to determine dispersal probability.
Under stable conditions, both architectures successfully reproduce classical patterns of density- and sex-dependent dispersal. However, a clear difference emerges once populations expand into new, empty territories. Evolved gene-regulatory networks harbour substantially more cryptic genetic variation, which is revealed under these changing conditions. Previously hidden variation becomes exposed to selection at the expanding front, where low-density conditions create novel selective pressures. As a result, dispersal increases significantly, accelerating range expansions compared to the simpler, single-locus architectures.
These findings highlight how the genetic architecture of ecologically relevant traits, such as dispersal, not only shapes range dynamics but can also influence how populations respond to the demographic and environmental shifts encountered during expansion. By demonstrating that gene-regulatory networks facilitate faster range expansions due to their ability to store and later reveal cryptic genetic variation, Deshpande and Fronhofer take a useful step toward integrating genetic complexity into eco-evolutionary models.
References
Ezoe, H. and Iwasa, Y. (1997), Evolution of condition-dependent dispersal: A genetic-algorithm search for the ESS reaction norm. Popul Ecol, 39: 127-137. https://doi.org/10.1007/BF02765258
Jhelam N. Deshpande, Emanuel A. Fronhofer (2023) A gene-regulatory network model for density-dependent and sex-biased dispersal evolution during range expansions. bioRxiv, ver.4 peer-reviewed and recommended by PCI Evol Biol