
Evolved gene-regulatory networks underlying dispersal plasticity can accelerate range expansions
 based on reviews by Arnaud Le Rouzic and 2 anonymous reviewers
based on reviews by Arnaud Le Rouzic and 2 anonymous reviewers
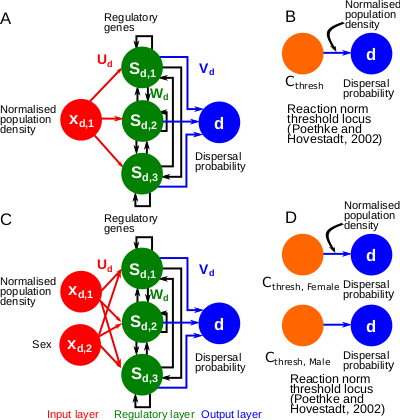
A gene-regulatory network model for density-dependent and sex-biased dispersal evolution during range expansions
Abstract
Recommendation: posted 15 March 2025, validated 18 March 2025
Mullon, C. (2025) Evolved gene-regulatory networks underlying dispersal plasticity can accelerate range expansions. Peer Community in Evolutionary Biology, 100749. 10.24072/pci.evolbiol.100749
Recommendation
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
This work was supported by a grant from the Agence Nationale de la Recherche (No.: ANR-19-CE02- 312 0015) to EAF.
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/2023.07.18.549508
Version of the preprint: 3
Author's Reply, 11 Mar 2025
Decision by Charles Mullon , posted 11 Mar 2025, validated 29 Sep 2024
, posted 11 Mar 2025, validated 29 Sep 2024
Dear authors,
Thank you once again for submitting the revised version of your manuscript, “A gene-regulatory network model for density-dependent and sex-biased dispersal evolution during range expansions,” for consideration at PCI Evol Biol. I appreciate the significant effort you’ve put into addressing the feedback from the previous review round. The improvements—particularly the rewritten introduction, new figures, and supplementary material—have certainly made the manuscript clearer.
After reviewing the revised manuscript and your responses, I decided to send it back to Reviewer 3, as I felt that some of their comments weren’t fully addressed. Their detailed feedback highlights several points that need further attention before we can move forward with a recommendation.
Reviewer 3 continues to find the comparison between the GRN and RN models difficult to interpret, and I agree. First, the way the two models are presented—RN as a solid line and GRN as a "cloud" of individual lines—makes it hard to directly compare them. They suggest using a more consistent format for both models (like showing average ± standard deviation or quartiles) to make things clearer.
The second concern, which I also share, is about the differences in mutational effects between the GRN and RN models. Supplementary figures (S4 and S8) show how mutation affects the reaction norms differently, but the reviewer mentions that these differences might come from the arbitrary settings of mutation effects, rather than being inherent to the GRN architecture. What needs to be clarified here is whether the larger standing genetic variation in the GRN model is truly a feature of its architecture or is influenced by parameter choices. Reviewer 3 suggests reducing the mutation size in the GRN model to bring it closer to the RN model, but you may have other ideas for how to address this.
There’s also a question about the apparent discrepancy between equilibrium dispersal rates and range expansion dynamics. In Figure 5A, dispersal is greater under the GRN model right from the first generation, but in Figure 3, the equilibrium dispersal rates between the two models seem pretty similar. Why don’t both models start at the same dispersal rate and then diverge over time as expected? Please clarify this point.
Please revise your manuscript to address these issues, along with the other points raised by Reviewer 3, which you’ll find in the detailed feedback below.
When you’re ready, please submit a revised manuscript along with a detailed, point-by-point response. Once these revisions are made, I’ll be able to continue with the evaluation and move toward a final decision.
Best regards,
Charles Mullon.
Reviewed by Arnaud Le Rouzic , 24 Sep 2024
, 24 Sep 2024
Review of "A gene-regulatory network model for density-dependent and sex-biased dispersal evolution during range expansions" by JN Deshpande and EA Fronhofer, PCI-EvolBiol-749.
This is a revised version of a ms submitted to PCI in December 2023. The ms has been read by three reviewers, which evaluation was (as far as I can tell) rather consistent. All three reviewers found some scientific merit to the manuscript and considered it to deserve publication; all also noticed some shortcomings that should be addressed.
The manuscript has been substantially re-written (up to the point that the "revision tracking" version is hard to follow). Large-scale changes (including new paragraphs, deleted paragraphs, and text restructuration) have been perfomed in all sections. Figure 1 has been added, and figures 2 and 3 have been redrawn with different colors and style. A series of supplementary figures have been added to address some of the reviewers' concerns.
I had 5 major comments on the previous version: 1) the introduction was lacking focus, 2) the rationale for contrasting with "first principle" predictions was unclear, 3) Model comparison was difficult from the figures (and I was unsure that both models were calibrated in terms of mutations), 4) Patterns from the range expansion figures did not seem to match the equilibrium dispersion values, 5) The population genetics of range expansion was not considered (e.g. selection/gene flow balance between the central and expanding patches). Some of these issues (especially 1 and 3) largely overlapped with other reviewers' concerns.
My concern #1 has been addressed for the most part. The authors have deeply rewritten the introduction, and I felt that is it now more to the point. It also refers more precisely to the relevant literature, and I think this concern has been mostly addressed.
My concern #2 has also been addressed. The authors have reframed the results so that the RN model is more clearly associated with a "reference" model, which is fine. Commenting more precisely on the parameter space in which the GRN model departs from the RN model (for instance in paragraph 221) convincingly clarifies the purpose of model comparison.
I am not sure that my concern #3 has been addressed in the revised version. I think all three reviewers shared the same feeling about the impossibility to compare models from figures 2 and 3. The authors have changed the figure design, but I am afraid that the new version is not better than the former one. The results from the RN model are displayed as a solid line standing for the reaction norm predicted from the median parameters (and interquartile predictions), while the results from the GRN model are individual simulated lines cumulated on the plot. As a result, both representations are so different that it is not possible to eyeball whether the predictions really match, beyond the fact that the purple line seems close to the "cloud" of green lines. The authors proposed to quantify the differences between models in supplementary figures, but as far as I understand, these represent the difference between both models and the mathematical optimum (i.e. not the difference between simulated RN and GRN).
Here, the authors decided to keep a visual representation that seemed to bother all three reviewers, and instead proposed supplementary figures, as if the point was to check some reviewers' concerns. I think it is natural, when the purpose is to comment on differences between model predictions, to propose figures in which models are treated in the same way. Both models predict a stationary distribution of reaction norms at equilibrium, and both could be computed and presented in an equivalent way (average +/- sd, median and quartiles, etc), so that it the difference could be eyeballed directly.
Note that the new fig 3 barely readable in my printed copy (color lines overlap and it seems impossible to eyeball the differences between GRN observations).
There might have also been some misunderstanding about my comment 3c. I totally agree that it is difficult to calibrate mutational effects across models, especially when models are so different. I think I also understand what the authors illustrate with sup figs S4 and S8: mutations not only have different effects in different models, they also affect the reaction norm differentially, so that their effect depends on the population density. In the GRN model, epistasis may also lead to the evolution of mutational effect, and one could thus expect that the effect of mutations may evolve even if the reaction norm is at equilibrium. Yet, this was exactly what I wanted to point out : because it is difficult to calibrate mutations across models, it is also difficult to conclude on potential differences in evolvability and speed of adaptation (standing genetic variation and mutational variance). Setting sigma_m = 1 leads to larger mutation size on dispersal in he GRN that in the RN, and this is arbitrary (the unit of sigma_m is different in both models). Even if I agree that increasing sigma in the RM model might not change the dispersal at low density, decreasing sigma in the GRN is likely to bring both models closer in terms of mutational effect on dispersal. As a consequence, the larger standing genetic variation in GRN simulations might be arbitrary to some extant. My point is that this might not only be a property of the GRN architecture, but rather a property of setting sigma_m to some specific values.
My concern #4 might have been unclear. In figures 4 and 5, the slope of the dispersal speed at t=0 is the dispersal rate at equilibrium. In what is now fig 5A (top left panel), the GRN model disperses faster from the first generation. However, in Fig 3 top left, there is not that much of a difference between equilibrium dispersal rates between RN and GRN (as far as I can see, but see comment #3 above about figure readability). I find it paradoxical, is evolution so fast that in fig 5 it is impossible to see that both populations start to disperse at the same rate before diverging?
My concern #5 was related to the fact that gene flow may affect adaptation, because the optimal dispersal rate for edge populations might be different than the more central populations. The authors consider that this is not central to the study, and I can hardly contradict them without simulations. From Figures 4 and 5, dispersion does not seem to evolve much (the expansion is rather linear), and gene flow from non-expanding populations could partly explain this observation. This concern #5 was not addressed, but it remains minor.
Minor comments: the authors have addressed most of them satisfactorily.
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2023.07.18.549508
Version of the preprint: 2
Author's Reply, 14 Aug 2024
Decision by Charles Mullon , posted 26 Feb 2024, validated 28 Feb 2024
, posted 26 Feb 2024, validated 28 Feb 2024
Dear authors,
First, I'd like to apologize for the time it has taken to provide you with feedback. Securing reviewers and navigating through the holiday season and personal commitments resulted in some delays. Nevertheless, I've managed to secure three expert reviews, which have provided valuable insights to improve your manuscript.
As you will discover from their comments, all three reviewers agree that your manuscript has merit but also important shortcomings. In particular, all agree that the introduction lacks focus and does not naturally converge to the questions addressed by the manuscript. They’ve also highlighted the need for a more exhaustive explanation of the model, particularly regarding the simulations' crux. I therefore encourage you to give more detail regarding the genotype-to-phenotype relationship and how mutations translate into phenotypic variation, as these aspects seem particularly relevant to understanding your results.
Reviewer 3 also raised concerns about several citations, and I echo their criticism. I additionally draw attention to the relevance of Ezoe, H., Iwasa, Y. Evolution of condition-dependent dispersal: A genetic-algorithm search for the ESS reaction norm. Res Popul Ecol 39, 127–137 (1997). This paper looks at the evolution of a dispersal reaction norm to local competition, and compares the results coming from an optimality mathematical argument and those from an evolutionary simulation of a neural network. There are therefore many similarities with the current study and it is imperative that you engage with this literature.
Additionally, the discussion on current theory of the evolution of reaction norms to social traits appears somewhat superficial (e.g., only one broad and non-technical review on adaptive dynamics is cited on l. 328). There are many mathematical results at our disposal to understand the evolution of reaction norms (see citations below for examples), and I think the Discussion section could do a better job at explaining how the use of GRN may help understand adaptation beyond these results.
I hope you'll consider addressing these comments in a revised manuscript. Please provide a point-by-point response to the reviewers' comments with your revision.
All the best,
Charles Mullon.
• Metz, J. A., & Diekmann, O. (1986). The dynamics of physiologically structured populations. Springer.
• Gomulkiewicz, R., & Kirkpatrick, M. (1992). Quantitative genetics and the evolution of reaction norms. Evolution, 46, 390-411.
• Avila, P., Priklopil, T., & Lehmann, L. (2021). Hamilton’s rule, gradual evolution, and the optimal (feedback) control of phenotypically plastic traits. Journal of Theoretical Biology, 526, 110602.
• Parvinen, K., Dieckmann, U., & Heino, M. (2006). Function-valued adaptive dynamics and the calculus of variations. Journal of Mathematical Biology, 52, 1-26.
• Durinx, M., Metz, J.A.J. & Meszéna, G. (2008). Adaptive dynamics for physiologically structured population models. J. Math. Biol. 56, 673–742.
• Gomulkiewicz, R., Kingsolver, J. G., Carter, P. A., & Heckman, N. (2018). Variation and evolution of function-valued traits. Annual Review of Ecology, Evolution, and Systematics, 49, 139-164.
Reviewed by Arnaud Le Rouzic , 01 Feb 2024
, 01 Feb 2024
Review of "A gene-regulatory network model for density-dependent and sex-biased dispersal evolution during range expansions" by J.N. Deshpande and A. Fronthofer, BioRxiv 2023.07.18.549508 for PCI Evol Biol #749.
This is a theoretical study focusing on the evolution of the plastic response of dispersal as a consequence of population density. Dispersal is a key life history trait for most organisms, as it allows the species to colonize new patches and to avoid extinction. It also promotes gene flow. Yet, knowing when and how much to disperse is a tricky question, as dispersal is generally associated with some risks, and leaving a comfortable patch for the unknown is not necessarily a good strategy. Furthermore, studying the evolution of dispersion-related genes is complicated, since the dispersal rate that maximizes the population survival is not necessarily the same as for the most successful allele. Here, the authors address the question of how the optimal dispersal strategy should depend on the population density; the benefits of dispersing being indeed much larger when competition is tough compared with the situation where many resources are available. The authors also address the question of optimal male vs female dispersal, and the consequences of a change in the availability of new patches.
The approach here is genuinely original. Instead of setting up a model where the plastic response (dispersal rate as a function of the population density) is a parameterized function, the authors assume that this behavioral trait results from the expression of several underlying genes, and simulate the evolution of a small gene network in a dynamic metapopulation setting. As a result, the plastic response is not expected to follow a pre-defined shape, which greatly limits the possibility that the model results are accidentally constrained by some modeling choices.
Here, the authors show that the plastic response of dispersal follows closely the theoretical expectation from the "RN" model by Poethke and Hoverstadt 2002: there is a density thershold below which dispersal should be avoided, and the optimal dispersal rate raises rapidly above the threshold before stabilizing. Yet, simulations also show the plastic response in the gene network simulations is featured by a large amount of cryptic genetic variation, which ensures a rapid genetic response when conditions change (typically, when new patches become abundant). The authors also found that, in their setting, males should disperse more than females, especially when the risk of local extinction is low.
Overall, there are several interesting points in this manuscript.
(i) using a flexible, but biologically realistic gene-network based genetic architecture to study the evolution of the shape of a reaction norm for a behavioral trait seems very relevant.
(ii) the studied trait (dispersal as a function of population density) has some non-trivial evolutionary properties, and deserves attention.
(iii) the conclusions of this work are compared with existing theoretical predictions, which is reassuring when models involve complex pieces of simulation software.
This study also has shortcomings. Some point that could be possibily fixed are listed below:
1. I am not completely sure that the introduction converges naturally towards the questions 1) and 2). I was rather unsure to understand what the paper was about before the very end of the introduction, and I think it would probably help to achieve faster a focus on specific questions. In addition, I found it difficult to find a natural connection between the different sub-questions (sex-specific behavior, shape of the reaction norm, and rate of expansion) beyond the fact that they are related to the evolution of dispersal.
2. I was confused for a long time (and I am still a bit unsure) about what the authors wanted to test exactly when comparing simulation results to the RN model. If the existing theoretical results are derived from first principles, any mismatch between these predictions and the simulations could only mean that the model implemented in the simulations did not match the assumptions of the mathematical model (unless there was a mistake when deriving this model). In other words, the RN model predicts "with certainty" the optimal reaction norm. There would be several reasons for the GRN model not to converge to the optimum; for instance, the genetic architecture might not be flexible enough to give this precise shape. Another possibility could be that Darwinian selection may not be powerful enough to precisely shape the plastic response (because some environments are too rare to induce enough selection pressure, or because of genetic correlations with other selected characters). Yet, any of these observations should not dismiss the RN model, because the model prediction about the optimal reaction norm would still be valid. A similar question arises with the sex-specific model; here, there is little doubt that sex-specific responses should arise in parallel in both GRN and RN models, what was the authors' expectations and what would have been the conclusion in case of mismatch?
3. A central point of this work is to compare the GRN predictions to the theoretical RN model, but the way results are presented does not make it easy to evaluate to what extent these results match.
3a: figures 2 and 3 display the mean +/- sd for the RN model, and individual phenotypes for the GRN model. I may have missed something, but I am not sure to understand why both are not represented in the same way. The full reaction norm could easily be computed for GRN genotypes (by changing the environment), or the simulated dispersal for the RN model could be restricted to the environment in which genotypes were sampled. Overall, it is indeed possible to eyeball that both results seem to match, but it is impossible to assess to what extent they do.
3b: the introduction mentions that alternative shapes for the plastic response were proposed in the literature. Even if the fit to the RN predictions seems convincing, I wonder if e.g. a sigmoid response would not be at least as convincing.
3c: a substantial part of the results is devoted to the difference between the GRN model and the RN predictions in terms of genetic variance in the population. Even if the authors make a convincing argument about the higher evolvability in the GRN architecture, I was not sure that the mutational variance in both models was calibrated. Indeed, how much single mutations affect the phenotype remains quite arbitrary in these models (mutational targets, such as W, V, or Cthresh, are not expressed in the same dimensional units). A larger effect of mutations would generate a larger mutational variance, and as a consequence, a larger genetic variance at equilibrium. Here, the authors state in Table 1 that mutations change the genotype with a standard deviation sigma_m, but it is unclear whether this sigma_m is the same across models. Does it mean that mutations affect W, V, and Cthresh with the same standard deviation? This could generate drastically different mutational variances on the dispersal itself, and thus different evolvabilities. I assume that it would be reasonable to calibrate the mutational effects in the different models before drawing conclusions on evolvability, by e.g. adjusting sigma_m so that the mutational variance of dispersal would be identical in some arbitrary standard conditions (e.g. d=0.5 at hat_N?).
4. In figures 4 and 5, the speed at which the species expand in the available patches depends on the genetic architecture. In many panels, the range expansion seems rather linear, with a different speed from the initial generation. As far as I understand, the authors' interpretation is that this is due to different evolvabilities of the dispersion reaction norm, but I do not understand why these differences do not always build up progressively (as in panels 4C or 4F, for instance). My interpretation of panels e.g., 5A and 5B is that the dispersion itself differs between models from generation 0, which does not seem to be the case in the corresponding panels 3A and 3B. Am I missing something here?
5. In the range expansion simulations, the genetic (and phenotypic) composition of populations probably varies as a function of time and distance from the origin. Figures 4 and 5 focus on the expansion, i.e. the alleles "surfing" the expansion wave, but what about the other parts of the landscape? Are central populations affected by the expansion? Does the increased dispersion at the edges of the species range generates some maladaptive gene flow towards saturated parts of the landscape?
A few minor issues:
* Introduction, lines 34 or 37 for instance, but at other places as well: the literature review at present tense ("They find that ...") sounds slightly confusing.
* Two first paragraphs of the introduction (up to line 64): the review is sometimes not very precise. For instance, it is said that some authors have studied or discussed the effect of different factors, but no indication about the conclusions of these studies.
* Line 100: is sexual dimorphism a special case of plasticity? According to the point of view, one could indeed consider that sexual dimorphism is just a consequence of the presence/absence of sex-specific hormones. However, in many organisms, sex determination is genetic, and sexual dimorphism would be a clear case of genetic determinism of the phenotype.
* Methods, line 129: What is the influence of the second (y) dimension of the landscape? If the same results were obtained with a one-dimensional landscape, is it really necessary to have the y dimension? And if y matters, why only 5 elements in the grid?
* I might have missed it, but I could not find the information that alpha and lambda_0 were chosen so that hat(N) = 100 (if my calculation is correct).
* In equation (3), the star is generally not considered as a correct mathematical symbol for multiplication.
* Figures 2 and 3, why are datapoints discretized on the x axis? If my understanding is correct, these data point should come with any value of x.
* Caption of figures 2 and 3: I am not sure that the acronym ES was defined before in the text (does it stand for environmental susceptibility?).
* Line 252: this sounds like discussion material, and not results.
* Line 266-274: I am not familiar with the problem of sex-related dispersal; I am quite convinced that the authors are correct but I confess I did not understand why dispersal was male-biased in the model. In particular, I was confused that from fig 3, it seems that male dispersion is quite constant across the different panels. Therefore, it seems more natural to say that females disperse less than males (especially when epsilon = 0), since the evolvable parameter seems to be female dispersal.
* The color/line code was completely different between figures 2/3 and 4/5, I found it slightly disturbing.
Reviewed by anonymous reviewer 1, 19 Jan 2024
Reviewed by anonymous reviewer 2, 23 Feb 2024
In this manuscript, the authors investigate the evolution of dispersal plasticity under equilibrium and range expansion regimes, with an individual-based meta-population model. To encode plasticity, they use a Wagner-like evolvable genetic regulation network (GRN) that senses the local population density and the sex of the individual. Through numerical simulations, the authors suggest that after long-term evolution at equilibrium (20,000 generations), GRNs evolved a plastic response similar to previous analytical predictions. During range expansion, GRNs allow a faster population dispersal than hard-encoded plastic responses.
I think that the authors addressed a very interesting question, by using a more complex genotype-to-phenotype map to evolve dispersal plasticity. However, there are several issues in the manuscript that need to be solved before considering it suitable for publication:
1) The structure and writing of the manuscript need improvement. The manuscript does not maintain a clear track of the question, and the reader can become lost in literature and general considerations that are sometimes out of context (see my comments below). Conciseness must also be improved.
2) The presentation of the model lacks clarity. Some features, such as population dynamics in each patch, sexual reproduction, and the mutational process, are poorly explained. I personally struggled to understand the detail of the model.
3) I also believe that the results are not presented in a convincing manner:
- At equilibrium, the comparison between GRN and RN relies solely on graphical interpretation. While this approach could be acceptable, commenting on hardly readable scatter plots is insufficient to demonstrate the similarity to RN curves. There are sometimes a large number of outliers, casting doubt on the results (e.g., the left column of Figs 2 and 3). The minimum requirement would be to plot a running mean/median/mode and perform some statistical or fitting tests to convince the reader.
- At range expansion, the authors extensively interpret the results without substantiating their claims with additional simulation insights. On this point, I believe that the mutation rate plays an important role, and the values used should be discussed, at least in the Methods section.
To conclude, I find the question and the methodology used interesting, and I believe the authors have the opportunity to improve their manuscript and strengthen their results with additional simulation insights (I am thinking about the male-biased dispersal and/or why GRNs spread faster than RNs). I certainly do not suggest re-running entire simulation campaigns but rather attempting to support some of the authors' claims with mechanistic simulation insights.
I believe that if the authors can address the issues mentioned above, this could make a valuable contribution to PCI EvolBiol.
Detailed comments:
==================
- General: check for spelling and conciseness.
Abstract:
---------
- Abstract (lines 2 -> 9): Improve the writing of this section. Should we distinguish evolution rate from plasticity evolution rate? What is the interplay?
- Abstract: Introduce one sentence to tell why GRN are biologically relevant, compared to RN (as extensively discussed in the introduction).
Introduction:
-------------
In general, prefer past form to present form, and focus more on the question (for example, it should be clear from the beginning that the GRN senses sex and/or population density, nothing more).
- l.22 remove "(evolution)" and "(ecology)", repetition of the previous line.
- l.26 remove "now".
- l.55, l.59: "fragmentation": I would avoid discussing this notion, as it is not explored in the manuscript.
- l. 73-89: shorten this section. Btw, if I am not wrong, this is not exactly a Wagner model (where the genotype is a single weight matrix).
- l. 91: "at the molecular level": a weight matrix is already a significant abstraction of molecular processes.
- l. 100: "organism's internal state": only individual sex is being sensed here.
- l. 101-112: the approach and bibliography are somewhat mixed up here.
- l. 113: "concretely": Avoid, as it should be clear to the reader already.
Model description:
------------------
- l. 122: focus on the question, the inputs are not external cues and internal states, but individual's sex and population density, i.e. two values.
- l. 137: "periodic" -> toroidal?
- l. 138: Does the simulation automatically stops when the border of the grid is reached?
- l. 145: "eight nearest neighboring patches": Moore neighborhood
- l. 152-159: Population dynamics are not clearly explained here.
- l. 160: "The parental generation then dies and is replaced by the offspring": Generations are non-overlapping.
- l.168-173: Could be removed (hence the "More concretely")
- l.178-188: Same problem here, too many general considerations.
- l. 194: "discard": how is it implemented in the simulation? Does the individual die?
- l. 217-218: "mu = 0.01, 0.1, 0.3" ==> mu \in {0.01, 0.1, 0.3} and elsewhere (\in LaTeX math symbol).
- l. 217: "dispersal costs" ==> dispersal mortality?
Results:
--------
- l. 222: "emerge from cellular and molecular processes": I would nuance with "can indeed emerge from a more complex genotype-to-phenotype map".
- l. 228: "Fig. 2 shows" ==> cf. to my main comment, the figure alone is not convincing enough.
- l. 231-235: not clear.
- Fig. 2: cf. my main comment. Especially for left-bottom panels containing GRN outliers.
- l. 246-256: perhaps this part should belong to the discussion.
- Fig. 3: same problem here. Moreover, it would be nice to have simulation mechanistic insights to better understand the male-biased dispersal.
- l. 266-274: perhaps this part should also belong to the discussion.
- Figs. 4 and 5: There is no legend to interpret the colors. Letters sometimes overlap with curves.
Discussion:
-----------
- l. 335: "robustness": I would avoid this notion and stay focused on the question.
- l. 344: "genotype-to-phenotype (GP)": this term is introduced too late in the manuscript.
- l. 346: "While empirical evidence supporting our work is scarce": another reason to seek more insights from the simulations.










