
Reticulated evolution marks the rapid diversification of the Delphinoidae
A genomic assessment of the marine-speciation paradox within the toothed whale superfamily Delphinoidea
Abstract
Recommendation: posted 01 July 2022, validated 04 July 2022
Fontaine, M. (2022) Reticulated evolution marks the rapid diversification of the Delphinoidae. Peer Community in Evolutionary Biology, 100139. https://doi.org/10.24072/pci.evolbiol.100139
Recommendation
Historically neglected or considered a rare aberration in animals under the biological species concept, interspecific hybridisation has by now been recognised to be taxonomically widespread, particularly in rapidly diversifying groups (Dagilis et al. 2021; Edelman & Mallet 2021; Mallet et al. 2016; Seehausen 2004). Yet the prevalence of introgressive hybridizations, its evolutionary significance, and its impact on species diversification remain a hot topic of research in evolutionary biology. The rapid increase in genomic resources now available for non-model species has significantly contributed to the detection of introgressive hybridization across taxa showing that reticulated evolution is far more common in the animal kingdom than historically considered. Yet, detecting it, quantifying its magnitude, and assessing its evolutionary significance remains a challenging endeavour with constantly evolving methodologies to better explore and exploit genomic data (Blair & Ané 2020; Degnan & Rosenberg 2009; Edelman & Mallet 2021; Hibbins & Hahn 2022).
In the marine realm, the dearth of geographic barriers and the large dispersal abilities of pelagic species like cetaceans have raised the questions of how populations and species can diverge and adapt to distinct ecological conditions in face of potentially large gene-flow, the so-called marine speciation paradox (Bierne et al. 2003). Contemporaneous hybridization among cetacean species has been widely documented in nature despite large phenotypic differences (Crossman et al. 2016). The historical prevalence of reticulated evolution, its evolutionary significance, and how it might have impacted the evolutionary history and diversification of the cetaceans have however remained elusive so far. Recent phylogenomic studies suggested that introgression has been prevalent in cetacean evolutionary history with instances reported among baleen whales (mysticetes) (Árnason et al. 2018) and among toothed whales (odontocetes), especially in the rapidly diversifying dolphins family of the Delphininae (Guo et al. 2021; Moura et al. 2020).
Analysing publicly available whole-genome data from nine cetacean species across three families of Delphinoidae – dolphins, porpoises, and monondontidae – using phylogenomics and demo-genetics approaches, Westbury and colleagues (2022) take a step further in documenting that evolution among these species has been far from a simple bifurcating tree. Instead, their study describes widespread occurrences of introgression among Delphinoidae, drawing a complex picture of reticulated evolutionary history. After describing major topology discordance in phylogenetic gene trees along the genome, the authors use a panel of approaches to disentangle introgression from incomplete lineage sorting (ILS), the two most common causes of tree topology discordances (Hibbins & Hahn 2022). Applying popular tests that separate introgression from ILS, such as the Patterson’s D (a.k.a. ABBA-BABA) test (Durand et al. 2011; Green et al. 2010), QuIBL (Edelman et al. 2019), and D-FOIL (Pease & Hahn 2015), the authors report that signals of introgression are present in the genomes of most (if not all) the cetacean species included in their study. However, this picture needs to be nuanced. Most introgression signals seem to echo old introgression events that occurred primarily among ancestors. This is suggested by the differential signals of topology discordance along the genome when considering sliding windows along the genome of varying sizes (50kb, 100kb, and 1Mb), and by patterns of excess derived allele sharing along branches of the species tree, as captured by the f-branch test (Malinsky et al. 2021; Malinsky et al. 2018). The authors further investigated the dynamic of cessation of gene flow (and/or ILS) between species using the F1 hybrid PSMC (or hPSMC) approach (Cahill et al. 2016). By estimating the cross-coalescent rates (CRR) between species pairs with time in light of previously estimated species divergence times (McGowen et al. 2020), the authors report that gene flow (and/or ILS) most likely has stopped by now among most species, but it may have lasted for more than half of the time since the species split from each other. According to the author, this result may reflect the slow process by which reproductive isolation would have evolved between cetacean lineages, and that species isolation was marked by significant introgression events.
Now, while the present study provides a good overview of how complex is the reticulated evolutionary history of the Delphinoidae, getting a complete picture will require overcoming a few important limitations. The first ones are methodological and related to the phylogenomic analyses. Given the sampling design with one diploid genome per species, the authors could not phase the data into the parental haplotypes, but instead relied on a majority consensus creating mosaic haploidized genomes representing a mixture between the two parental copies. Moreover, by using large genomic windows (≥50kb) that likely span multiple independent loci, phylogenetic analyses in windows encompassed distinct phylogenetic signals, potentially leading to bias and inaccuracy in the inferences. Thawornwattana et al (2018) previously showed that this “concatenation approach” could significantly impact phylogenetic inferences. They proposed instead to use loci small enough to minimise the risk of intra-locus recombination and to consider them in blocks of non-recombining loci along the genome in order to conduct phylogenetic analysed, ideally under the multi-species coalescent (MSC) that can account for ILS (e.g. BPP; Flouri et al. 2018; Jiao et al. 2020; Yang 2015). Such an approach applied to the diversification of the Delphinidae may reveal substantial changes compared to the currently admitted species tree.
Inaccuracy in the species tree estimation may have a major impact on the introgression analyses conducted in this study since the species tree and branching order must be supplied in the introgression analyses to properly disentangle introgression from ILS. Here, the authors rely on the tree topology that was previously estimated in McGowen et al. (2020), which they also recovered using their consensus estimation from ASTRAL-III (Zhang et al. 2018). While the methodologies accounted to a certain extent for ILS, albeit with potential bias induced by the concatenation approach, they ignore the presumably large amount of introgression among species during the diversification process. Estimating species branching order while ignoring introgression can lead to major bias in the phylogenetic inference and can lead to incorrect topologies. Even if these MSC-based methods account for ILS, inferences can become very inaccurate or even break down as gene flow increases (see for ex. Jiao et al. 2020; Müller et al. in press; Solís-Lemus et al. 2016). Dedicated approaches have been developed to model explicitly introgression together with ILS to estimate phylogenetic networks (Blair & Ané 2020; Rabier et al. 2021) or in isolation-with-migration model (Müller et al. in press; Wang et al. 2020). Future studies revisiting the reticulated evolutionary history of the Delphinoidae with such approaches may not only precise the species branching order, but also deliver a finer view of the magnitude and prevalence of introgression during the evolutionary history of these species.
A final part of Westbury et al. (2022)'s study set out to test whether historical periods of low abundance could have facilitated hybridization among Delphinoidae species. During these periods of low abundance, species may encounter only a limited number of conspecifics and may consider individuals from other species as suitable mating partners, leading to hybridisation (Crossman et al. 2016; Edwards et al. 2011; Westbury et al. 2019). The authors tested this hypothesis by considering genome-wide genetic diversity (or heterozygosity) as a proxy of historical effective population size (Ne), itself as a proxy of the evolution of census size with time. They also try to link historical Ne variation (from PSMC, Li & Durbin 2011) with their estimated time to cessation of gene flow or ILS (from the CRR of hPSMC). However, no straightforward relationship was found between the genetic diversity and the propensity of species to hybridize, nor was there any clear link between Ne variation through time and the cessation of gene flow or ILS. Such a lack of relationship may not come as a surprise, since the determinants of genome-wide genetic diversity and its variation through evolutionary time-scale are far more diverse and complex than just a direct link with hybridization, introgression, or even with the census population size. In fact, genetic diversity results from the balance between all the evolutionary processes at play in the species' evolutionary history (see the review of Ellegren & Galtier 2016). Other important factors can strongly impact genetic diversity, including demography and structure, but also linked selection (Boitard et al. 2022; Buffalo 2021; Ellegren & Galtier 2016).
All in all, Westbury and coll. (2022) present here an interesting study providing an additional step towards resolving and understanding the complex evolutionary history of the Delphinoidae, and shedding light on the importance of introgression during the diversification of these cetacean species. Prospective work improving upon the taxonomic sampling, with additional genomic data for each species, considered with dedicated approaches tailored at estimating species tree or network while accounting for ILS and introgression will be key for refining the picture depicted in this study. Down the road, altogether these studies will contribute to assessing the evolutionary significance of introgression on the diversification of Delphinoides, and more generally on the diversification of cetacean species, which still remain an open and exciting perspective.
References
Árnason Ú, Lammers F, Kumar V, Nilsson MA, Janke A (2018) Whole-genome sequencing of the blue whale and other rorquals finds signatures for introgressive gene flow. Science Advances 4, eaap9873. https://doi.org/10.1126/sciadv.aap9873
Bierne N, Bonhomme F, David P (2003) Habitat preference and the marine-speciation paradox. Proceedings of the Royal Society of London. Series B: Biological Sciences 270, 1399-1406. https://doi.org/10.1098/rspb.2003.2404
Blair C, Ané C (2020) Phylogenetic Trees and Networks Can Serve as Powerful and Complementary Approaches for Analysis of Genomic Data. Systematic Biology 69, 593-601. https://doi.org/10.1093/sysbio/syz056
Boitard S, Arredondo A, Chikhi L, Mazet O (2022) Heterogeneity in effective size across the genome: effects on the inverse instantaneous coalescence rate (IICR) and implications for demographic inference under linked selection. Genetics 220, iyac008. https://doi.org/10.1093/genetics/iyac008
Buffalo V (2021) Quantifying the relationship between genetic diversity and population size suggests natural selection cannot explain Lewontin's Paradox. e-Life 10, e67509. https://doi.org/10.7554/eLife.67509
Cahill JA, Soares AE, Green RE, Shapiro B (2016) Inferring species divergence times using pairwise sequential Markovian coalescent modelling and low-coverage genomic data. Philos Trans R Soc Lond B Biol Sci 371, 20150138. https://doi.org/10.1098/rstb.2015.0138
Crossman CA, Taylor EB, Barrett‐Lennard LG (2016) Hybridization in the Cetacea: widespread occurrence and associated morphological, behavioral, and ecological factors. Ecology and Evolution 6, 1293-1303. https://doi.org/10.1002/ece3.1913
Dagilis AJ, Peede D, Coughlan JM, Jofre GI, D’Agostino ERR, Mavengere H, Tate AD, Matute DR (2021) 15 years of introgression studies: quantifying gene flow across Eukaryotes. biorXiv, 2021.1106.1115.448399. https://doi.org/10.1101/2021.06.15.448399
Degnan JH, Rosenberg NA (2009) Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol 24, 332-340. https://doi.org/10.1016/j.tree.2009.01.009
Durand EY, Patterson N, Reich D, Slatkin M (2011) Testing for ancient admixture between closely related populations. Mol Biol Evol 28, 2239-2252. https://doi.org/10.1093/molbev/msr048
Edelman NB, Frandsen PB, Miyagi M, Clavijo B, Davey J, Dikow RB, Garcia-Accinelli G, Van Belleghem SM, Patterson N, Neafsey DE, Challis R, Kumar S, Moreira GRP, Salazar C, Chouteau M, Counterman BA, Papa R, Blaxter M, Reed RD, Dasmahapatra KK, Kronforst M, Joron M, Jiggins CD, McMillan WO, Di Palma F, Blumberg AJ, Wakeley J, Jaffe D, Mallet J (2019) Genomic architecture and introgression shape a butterfly radiation. Science 366, 594-599. https://doi.org/10.1126/science.aaw2090
Edelman NB, Mallet J (2021) Prevalence and Adaptive Impact of Introgression. Annual Review of Genetics 55, 265-283. https://doi.org/10.1146/annurev-genet-021821-020805
Edwards CJ, Suchard MA, Lemey P, Welch JJ, Barnes I, Fulton TL, Barnett R, O'Connell TC, Coxon P, Monaghan N, Valdiosera CE, Lorenzen ED, Willerslev E, Baryshnikov GF, Rambaut A, Thomas MG, Bradley DG, Shapiro B (2011) Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr Biol 21, 1251-1258. https://doi.org/10.1016/j.cub.2011.05.058
Ellegren H, Galtier N (2016) Determinants of genetic diversity. Nat Rev Genet 17, 422-433. https://doi.org/10.1038/nrg.2016.58
Flouri T, Jiao X, Rannala B, Yang Z (2018) Species Tree Inference with BPP Using Genomic Sequences and the Multispecies Coalescent. Mol Biol Evol 35, 2585-2593. https://doi.org/10.1093/molbev/msy147
Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, Hansen NF, Durand EY, Malaspinas AS, Jensen JD, Marques-Bonet T, Alkan C, Prufer K, Meyer M, Burbano HA, Good JM, Schultz R, Aximu-Petri A, Butthof A, Hober B, Hoffner B, Siegemund M, Weihmann A, Nusbaum C, Lander ES, Russ C, Novod N, Affourtit J, Egholm M, Verna C, Rudan P, Brajkovic D, Kucan Z, Gusic I, Doronichev VB, Golovanova LV, Lalueza-Fox C, de la Rasilla M, Fortea J, Rosas A, Schmitz RW, Johnson PLF, Eichler EE, Falush D, Birney E, Mullikin JC, Slatkin M, Nielsen R, Kelso J, Lachmann M, Reich D, Paabo S (2010) A draft sequence of the Neandertal genome. Science 328, 710-722. https://doi.org/10.1126/science.1188021
Guo W, Sun D, Cao Y, Xiao L, Huang X, Ren W, Xu S, Yang G (2021) Extensive Interspecific Gene Flow Shaped Complex Evolutionary History and Underestimated Species Diversity in Rapidly Radiated Dolphins. Journal of Mammalian Evolution 29, 353-367. https://doi.org/10.1007/s10914-021-09581-6
Hibbins MS, Hahn MW (2022) Phylogenomic approaches to detecting and characterizing introgression. Genetics 220, iyab173. https://doi.org/10.1093/genetics/iyab173
Jiao X, Flouri T, Rannala B, Yang Z (2020) The Impact of Cross-Species Gene Flow on Species Tree Estimation. Syst Biol 69, 830-847. https://doi.org/10.1093/sysbio/syaa001
Li H, Durbin R (2011) Inference of human population history from individual whole-genome sequences. Nature 475, 493-496. https://doi.org/10.1038/nature10231
Malinsky M, Matschiner M, Svardal H (2021) Dsuite - Fast D-statistics and related admixture evidence from VCF files. Mol Ecol Resour 21, 584-595. https://doi.org/10.1111/1755-0998.13265
Malinsky M, Svardal H, Tyers AM, Miska EA, Genner MJ, Turner GF, Durbin R (2018) Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nature Ecology & Evolution 2, 1940-1955. https://doi.org/10.1038/s41559-018-0717-x
Mallet J, Besansky N, Hahn MW (2016) How reticulated are species? Bioessays 38, 140-149. https://doi.org/10.1002/bies.201500149
McGowen MR, Tsagkogeorga G, Alvarez-Carretero S, Dos Reis M, Struebig M, Deaville R, Jepson PD, Jarman S, Polanowski A, Morin PA, Rossiter SJ (2020) Phylogenomic Resolution of the Cetacean Tree of Life Using Target Sequence Capture. Syst Biol 69, 479-501. https://doi.org/10.1093/sysbio/syz068
Moura AE, Shreves K, Pilot M, Andrews KR, Moore DM, Kishida T, Möller L, Natoli A, Gaspari S, McGowen M, Chen I, Gray H, Gore M, Culloch RM, Kiani MS, Willson MS, Bulushi A, Collins T, Baldwin R, Willson A, Minton G, Ponnampalam L, Hoelzel AR (2020) Phylogenomics of the genus Tursiops and closely related Delphininae reveals extensive reticulation among lineages and provides inference about eco-evolutionary drivers. Molecular Phylogenetics and Evolution 146,107047. https://doi.org/10.1016/j.ympev.2020.106756
Müller NF, Ogilvie HA, Zhang C, Fontaine MC, Amaya-Romero JE, Drummond AJ, Stadler T (in press) Joint inference of species histories and gene flow. Syst Biol.
Pease JB, Hahn MW (2015) Detection and Polarization of Introgression in a Five-Taxon Phylogeny. Syst Biol 64, 651-662. https://doi.org/10.1093/sysbio/syv023
Rabier CE, Berry V, Stoltz M, Santos JD, Wang W, Glaszmann JC, Pardi F, Scornavacca C (2021) On the inference of complex phylogenetic networks by Markov Chain Monte-Carlo. PLoS Comput Biol 17, e1008380. https://doi.org/10.1371/journal.pcbi.1008380
Seehausen O (2004) Hybridization and adaptive radiation. Trends Ecol Evol 19, 198-207. https://doi.org/10.1016/j.tree.2004.01.003
Solís-Lemus C, Yang M, Ané C (2016) Inconsistency of Species Tree Methods under Gene Flow. Syst Biol 65, 843-851. https://doi.org/10.1093/sysbio/syw030
Thawornwattana Y, Dalquen D, Yang Z, Tamura K (2018) Coalescent Analysis of Phylogenomic Data Confidently Resolves the Species Relationships in the Anopheles gambiae Species Complex. Molecular Biology and Evolution 35, 2512-2527. https://doi.org/10.1093/molbev/msy158
Wang K, Mathieson I, O’Connell J, Schiffels S (2020) Tracking human population structure through time from whole genome sequences. PLOS Genetics 16, e1008552. https://doi.org/10.1371/journal.pgen.1008552
Westbury MV, Cabrera AA, Rey-Iglesia A, Cahsan BD, Duchêne DA, Hartmann S, Lorenzen ED (2022) A genomic assessment of the marine-speciation paradox within the toothed whale superfamily Delphinoidea. bioRxiv, 2020.10.23.352286, ver. 7 peer-reviewed and recommended by Peer Community in Evolutionary Biology. https://doi.org/10.1101/2020.10.23.352286
Westbury MV, Petersen B, Lorenzen ED (2019) Genomic analyses reveal an absence of contemporary introgressive admixture between fin whales and blue whales, despite known hybrids. PLoS ONE 14, e0222004. https://doi.org/10.1371/journal.pone.0222004
Yang Z (2015) The BPP program for species tree estimation and species delimitation. Current Zoology 61, 854-865. https://doi.org/10.1093/czoolo/61.5.854
Zhang C, Rabiee M, Sayyari E, Mirarab S (2018) ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19, 153. https://doi.org/10.1186/s12859-018-2129-y
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
The work was supported by the Independent Research Fund Denmark | Natural Sciences, Forskningsprojekt 1, grant no. 8021-00218B and the Villum Fonden Young Investigator Programme, grant no. 13151 to EDL. AAC was funded by the Rubicon-NWO grant (project 019.183EN.005)
Evaluation round #5
DOI or URL of the preprint: https://doi.org/10.1101/2020.10.23.352286
Version of the preprint: v5
Author's Reply, 28 Mar 2022
Decision by Michael C. Fontaine, posted 09 Mar 2022
Dear authors,
Thank very much for having revised and resubmitted your manuscript. I have conducted a carrefull reading of this new version of your manuscrip and I do still have a few questions, issues and points that need to be adressed before I can accept to recommend your manuscript.
I have incorporated my comments and suggestions directly as comments into the pdf.
I hope this is ok with you.
I am staying at your disposal if you have any questions.
I am really looking forward reading your revised version and have the pleasure to recommend it.
Best wishes
Michael C FONTAINE
Download recommender's annotationsEvaluation round #4
DOI or URL of the preprint: https://doi.org/10.1101/2020.10.23.352286
Version of the preprint: v4
Author's Reply, 25 Feb 2022
Decision by Michael C. Fontaine, posted 16 Dec 2021
Dear Dr. Westbury and colleagues,
We have now received the reviewer's report for your manuscript. While like me he found that this new version has been improved a lot, and the new analyses you added have brought some clarity, the text is still in need of some important clarifications. The reviewer has raised six points to address raised, and I agree with him. Thus, I am inviting you to revise your manuscript considering carefully the reviewer’s comments, and resubmit a revised version.
At this point, I consider this as a minor, but important revision.
I look forward to receiving your revised manuscript.
Best Wishes,
Michael C FONTAINE
Reviewed by Simon Henry Martin, 09 Dec 2021
The authors have revised their manuscript, addressing my requests for more interpretation of the introgression analyses, and to add the f-branch analysis. I think brings some clarity to the results, suggesting that ancestral gene flow might be responsible for many of the observed signals. Having said that, there is no easy way to interpret all of the results, and I think this paper might highlight how at some level of complexity, different introgression scenarios become impossible to accurately reconstruct. I would not suggest any further analyses for this paper, but I do have some more suggestions to clarify the message/reasoning.
1. Despite the extensive evidence for gene flow on a per species level, it is important to note that the estimated proportions of the genome affected are small. Looking at the fb estimates of Malinsky et al. 2018, many are over 5%, whereas none of the estimates in the present study reach that level, with most below 1%. This suggests widespread gene flow but in small quantities, though it is also worth noting that f estimates tend to be lower the older the introgression event (Martin et al. 2015 MBE).
2. I generally agree with the interpretation of the QuIBLE results in tables S3 and S4, described in lines 149-168. However, both the text and table seem to use the term “outgroup” incorrectly. For example, in the triplet ((pilot, bot), orca), it is not correct to refer to pilot as the outgroup. I don’t know what the ideal term would be. In a hypothesis-based framework such as ((African, European), Neanderthal), it may be suitable to use the term “control taxon”, but since there is no prediction that one ingroup would be more likely to be subject to gene flow than the other, this seems wrong. Perhaps just the term “other ingroup” would be acceptable.
3. Line 312:
“When considering the uppermost limit of when two target genomes coalesce (equating the oldest date), and the lower confidence interval of each divergence date (equating the most recent date) (McGowen et al., 2020)”
I think this statement is talking about comparing the lower bound of the divergence time with the upper bound of the estimated cessation of gene flow, but the way it’s worded doesn’t sound like that to me.
4. hPSMC and ILS
Various lines seem to be suggesting that the hPSMC estimate of the date of cessation of gene flow could also indicate ILS of ancestral alleles in the absence of gene flow (line 315, 321, 324, 336, 340, 342, 345). My understanding is that the hPSMC approach used gives an estimate of when gene flow ceased, not when lineage sorting ceased. Incompletely sorted alleles could persist after the cessation of gene flow, and their coalescence times will necessarily all be older than the cessation of gene flow, so I don’t think the hPSMC result tells us anything about lineage sorting.
Could the hPSMC cessation of gene flow estimates instead be biased downward by some artefact? One idea that comes to mind is that mapping to the baji genome could bias the analysis to conserved regions, leading to underestimated coalescence time estimates.
5. Node age analysis and ILS
In their analysis of node ages as additional evidence for recent gene flow, the authors again seem to suggest that lineage sorting can result in recent node estimates (line 396). I don’t think this is right. If a gene tree groups killer whale with white-sided dolphin (Figure S6 B) due to ancestral ILS alone, the node would be older than the speciation date, not younger. The authors seem to agree with this in line 407. So I think the authors can confidently attribute this younger date to gene flow.
Line 408-409 seems to then show some misunderstanding. A gene tree that is discordant with the species tree can arise through ILS in the ancestral population, and does not require that ILS persisted after the speciation event. The gene tree can forever remain discordant with the species tree after lineage sorting is complete.
6. Finally, just a comment:
The hypothesis that many of the apparent species-specific gene flow signals could indicate differential retention of ancestrally introgressed tracts in descendent lineages is interesting, and should be considered in future studies. Assuming the gene flow occurred more than just a few generations prior to the diversification of each family, we would expect there to be many small introgressed tracts in the genome at the time of speciation, with a similar number of small introgressed tracts in each individual. Therefore, under neutrality, we would not expect the variance in introgressed proportions to change very much after the diversification of each family. To me, this leads to two conclusions. First, much of the introgressed alleles had not reached fixation or loss by the time the families diversified (otherwise in would be impossible to purge the introgressed tracts). Second, selection must have been involved in purging introgressed variation from some lineages more than others to create the significant differences observed.
https://doi.org/10.24072/pci.evolbiol.100359.rev41
Evaluation round #3
DOI or URL of the preprint: https://doi.org/10.1101/2020.10.23.352286
Author's Reply, 12 Nov 2021
Decision by Michael C. Fontaine, posted 13 Sep 2021
Dear Dr. Westbury and colleagues,
After reading again the manuscript in light of the previous rounds of revision, I am still convinced that this work is of great interest, but there are still some issues remaining that need to be addressed.
I have decided to ask for a fresh look from an expert in the field of reticulated evolution, Dr. Simon Martin and I have provided also detailed feedbacks incorporated in the attached version of the manuscript. Dr. Martin’s review and feedbacks encapsulate many of the remarks and issues I have on this manuscript, but I do have also some specifics of my own (in green in the pdf). Please check the reviewer’s feedbacks/comments and mine in the annotated pdf version.
I would advise the authors to follow these suggestions. I think they would benefit greatly to the manuscript and make it clearer and more accessible.
I am looking forward to receiving the revised version in order to complete the recommendation in the PCI Evol Biol.
Best regards
Michael C. FONTAINE
Download recommender's annotationsReviewed by Simon Henry Martin, 31 Aug 2021
Westbury et al. present a genome-scale assessment of gene flow in the Delphinoidea. Using a combination of approaches that consider genealogical discordance and branch length, they find evidence for a strikingly abundant “post divergence” gene flow in this group, including between families. While I initially found the results hard to believe, I do think that the most parsimonious interpretation of these findings is that the history of this group is far from a simple bifurcating process, due (at least in part) to widespread gene flow causing systematic discordance in genealogies across the genome.
Overall, I think this study is a good first step towards understanding speciation and the role of hybridisation in this lineage. Nevertheless, I still have multiple suggestions for improvements to the Results and Discussion section, as I felt that it was too brief and did not adequately describe the findings and their possible interpretations. I also have one suggestion for an additional analysis that I think will help with interpretation.
1. QuIBL
The results from the QuIBL analyses are described with a single vague sentence “Our QuIBL analyses suggest that the different retrieved topologies cannot be explained by ILS alone, but a combination of both ILS and gene flow.”
This is quite unsatisfactory as a description for such a broad set of tests. I would recommend adding a more detailed description of which pairs of non-sister taxa showed evidence for gene flow. Looking at tables S3 and S4, the results are striking and not immediately easy to interpret. There are so many significant signals, including support for gene flow between killer whales and every other member of the Delphinidae! I think the wording in the main text needs to capture at least the scale of significant signals, and ideally some discussion of what this would imply if the results are accurate (i.e. that gene flow might be or have been rampant between these species).
I also think the description of supplementary tables S3 and S4 requires more detail, as some of the column headings are not clear to the reader who does not know the QuIBL program.
2. D statistics
Like the QuIBL results, the D statistic results are not actually described in the manuscript, except a general statement that “85 out of 86 tests show signs of gene flow” (line 146). Looking at Table S5, we see that the results are even more astounding than those of QuIBL. Again, I recommend adding some description and interpretation in the text. Which pairs are consistently showing evidence for gene flow? Are the results consistent with QuIBL? Could any of the results indicate some systematic problem like the tree being incorrect?
It is good that the authors acknowledge that ancient gene flow events can be difficult to distinguish from multiple more recent events. However, they do not offer any detail as to how and where ancient gene flow might explain their specific findings in this study. For example, could ancient gene flow between killer whale and the ancestor of some dolphin species explain some of the apparent signals better than multiple more recent events? I know the authors have already added several additional analyses to appease reviewers, but one more I would strongly recommend is the f-branch statistic (Malinsky et al. 2018 Nature Ecology and Evolution, https://github.com/millanek/Dsuite) which provides an explicit approach to identify ancient events that affect the descendent lineages.
3. D-foil
The description of these results is better, as it provides some interpretation and possible caveats. However, the final statement “we suggest our result reflects the limited ability of D-foil to infer gene flow between these highly divergent lineages.” needs more explanation.
4. hPSMC
The hPSMC results suggest that gene flow continued until within the past 5 million years between all species considered. These times are difficult to reconcile with the divergence times estimated by McGowan et al. 2020, which are between 10 and 20 million years for most species. The authors appear to interpret the McGowan estimate as the “divergence time” and the hPSMC estimate as the end of “post-divergence gene flow”, but I think this reasoning needs to be justified by explaining why the McGowan estimate might be less impacted by post-divergence gene flow.
5. Tree node age analysis
The node age analysis is a nice addition to the paper, but the authors should acknowledge that it is not really independent from QuIBL. Arguably, QuIBL does a better job because it models the expected mixture of two different node age distributions resulting from ILS and gene flow. Nonetheless, the tree-based approach has appeal due to its simplicity, and the fact that we can consider multiple nodes at once.
I would also like to see some explanation for the choice of the three focal species used. Specifically, why were the two bottlenose species not included despite also showing ample evidence for gene flow in the other analyses?
6. Geography
There is quite a bit of text about allopatric vs parapatric speciation, but there is nothing about the distributions of these species, how much they overlap, and whether parapatry/allopatry is/was likely. Some information about their distributions will aid interpretation and could also help set up some predictions at the start of the paper, which would make it less descriptive.
https://doi.org/10.24072/pci.evolbiol.100359.rev31
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/2020.10.23.352286
Author's Reply, 28 Jun 2021
Decision by Michael C. Fontaine, posted 15 May 2021
Dear authors,
The manuscript has now gone through a second round of revision by the same three reviewers as the first round. All the reviewers have stated again the great merits of this study, and I share this point of view. All also felt a bit disapointed that the authors did not tried (or were not able to try) an alternative method to hPSMC that can handle both complete lineage sorting AND gene flow at the same time and test how comparable results are. Reviewer #1 (Christelle Fraisse, CF) suggested to try MSMC-IM even on a subset of the species for which suitable data would be available, and test how comparable would be the results from hPSMC and this methods. If this is not possible, may I suggest to try the new Approximate IM model of Müller et al. (Joint inference of species histories and gene flow. bioRxiv 348391; doi: https://doi.org/10.1101/348391) which can handle one single reference genome per species and infer jointly the species tree, the split time among species, and gene flow among them while accounting for ILS and gene flow. Either way could really strenghten the results, the scope, and the conclusions of this study and adress at least in part the reserves raised by reviewer #1 (CF) and reviewer 3 (by Andy Foote).
Beside that, some minor issues were pointed by reviewer #1 (CF) that can be fixed easily.
Assuming the authors would be able to validate the results of hPSMC with an independent approach, this would become a very strong study that I would be proud and honored to recommend.
I am staying at the authors disposal if they have any further questions and comments.
All the best
Michael C. FONTAINE
Recommender at PCI Evol Biol
Reviewed by Christelle Fraïsse , 08 Apr 2021
, 08 Apr 2021
The authors made efforts to address most of the concerns raised during the reviewing process, which is a positive point. Especially, they:
i) added a new tree topology-based approach (QuIBL).
ii) tested the robustness of the tree topology-based approach to GC content.
iii) tested the robustness of the hPSMC approach to repeat regions.
iv) they did bootstraps of the hPSMC for some species pairs.
Despite these great additions, I was left a bit disappointed that the authors did not compare their hPSMC results with that of MSMC-IM. It would have been a way to validate the timings for the cessation of gene flow with a second method. Given the various concerns that all reviewers raised with the hPSMC method, and because the occurrence of post-divergence gene flow long after initial divergence is a crucial point made by the authors, I think it is important to consolidate this part of the manuscript. The authors justified not using MSMC-IM because they “do not have access to phased data and/or population-level data for all of our species to be able to implement these analyses in a meaningful way so as to be able to make comparisons within the superfamily as is the focus of the study.” In my opinion, testing the MSMC-IM method on a subset of species will already be informative to assess if the timings of cessation of gene flow are congruent between methods.
Apart from this point, I am very satisfied with the revisions made.
Minor comments on the figures:
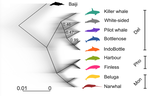
●Figure 1: some of the cartoons are shifted, and so they do not correspond to the correct branch. Also, the grey color gradient is hard to visualize. Why not simply using the same color code as in Figure 2A.
●Figure 2: please add “divergence time” to the right of the corresponding symbol in the legend.
https://doi.org/10.24072/pci.evolbiol.100359.rev21
Reviewed by anonymous reviewer 2, 05 May 2021
First and foremost, I apologize to the authors for my slowness in turning in the two reviews for each of the two rounds. My general opinion is that the article presented here has several interests. First, it focuses on gene flow over a broad evolutionary scale in a group of marine mammals. Allowing in the future to shed light on possible elements involved in the marine-speciation paradox by comparing with terrestrial mammals.
A second interest is that it can serve as a methodological textbook for this type of data in clades where population data are not available. A third interest is that it raises a demographic curiosity with such long periods of genetic exchange in time that are still to be understood.
Of course, I'm still not fully convinced by the title. Speciation "in the face of gene flow" suggests that barriers between species have evolved in spite of gene flow. This is different from having gene flow in spite of pre-established barriers. But the authors have refined this message in the text, which is essential. In the same way, the authors considered some suggestions made, including checking for possible biases related to GC content. This is an effort on their part that should be highlighted.
I am still struck by some of the quantified results, especially the strong support of gene flow, within families, over several million years. On a personnal note, I am really looking forward to seeing results using other inferential methods when data in multiple individuals will be available.
Thus, after this second round of revision, I have no further comments to make and can only support the current manuscript.
Camille Roux
https://doi.org/10.24072/pci.evolbiol.100359.rev22Reviewed by Andrew Foote , 06 Apr 2021
The authors have made some updates to their manuscript in response to the previous reviews and have added an additional analyses, based on measuring branch length variation along the genome. The conclusion drawn, that 'Speciation in the face of gene flow within the toothed whale superfamily Delphinoidea' still largely hangs on the author's interpretation of the hPSMC method.
I don't share the author's confidence that this method differentiates between gene-flow and lineage sorting. Essentially the upsweep in estimated Ne in the pseudo-diploid PSMC plots, which the authors infer as a cessation of gene flow, reflects a change in coalescence. And the inference that gene-flow continued after speciation was initiated is drawn from the comparison of the timing of the change in the trajectory of the hPSMC plot compared with the split time estimated by McGowen et al. The mapping of short read data to the distantly related baiji may be favouring the mapping to more conserved regions and may be a factor in the disagreement between the divergence estimates by McGowen et al. and the coarse estimate of the timing changes in coalescence interpreted from the PSMC plots.
I would recommend the authors to be more conservative in their interpretation of the PSMC plots of unphased pseudo-diploid genome generated by mapping short-reads to a distant relative as gene flow events that occurred millions of years ago.
https://doi.org/10.24072/pci.evolbiol.100359.rev23
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2020.10.23.352286
Author's Reply, 18 Feb 2021
Decision by Michael C. Fontaine, posted 30 Dec 2020
Dear authors,
The manuscript entitled “Speciation in the face of gene flow within the toothed whale superfamily Delphinoidea” by Westbury and colleagues has been now evaluated by three referees. They all agree that this study has great merit and should be of interest to the community. However, the three reviewers also raised major issues and concerns that I am also sharing.
In particular, the three reviewers acknowledged that the phylogenetic analyses were properly conducted, the interpretation from the results were sounds and the limitations properly acknowledged. See however the comments made by the three reviewers, especially reviewer 1 about incongruences in tree topologies compared to the consensus tree, which I am also sharing. The authors should also check the recently develop approach "QUIBL" developed by Edelmen and colleagues (2019; Science: 10.1126/science.aaw2090) to test for introgression without the limitation of symmetric tree topology imposed to the DFOIL test. This would be complementary to the test currently presented in the manuscript.
Regarding the second part of the analyses which aims at assessing and modeling post-divergence gene flow based on the use of TMRCA variation along the genome inferred from hPSMC, the three reviewers raised serious concerns and all agreed this part need improvement and I agree with them. This is especially important, since the title relies strongly on this part, and yet the results supporting this "speciation with gene flow" deserve further attentions and validations. The three reviewers made very good suggestions to explore the data further, interpret the results and discuss better their limitations. These would improve the study greatly.
I strongly suggest the authors to consider all the reviewers' recommendations.
I would be happy to reconsider this manuscript for a recommendation in PCI Evol Biol if the authors can continue to develop this study further and address point by point the three reviewer's comments.
Best wishes and already Happy New Year.
Michael C FONTAINE
Ref. Edelman N. B., et al. 2019 Genomic architecture and introgression shape a butterfly radiation. Science 366: 594–599.
Reviewed by Christelle Fraïsse , 26 Nov 2020
, 26 Nov 2020
Westbury and collaborators present an interesting study investigating post-divergence gene-flow in toothed whales. I liked the multifaceted approach combining phylogenetics, descriptive summary statistics and demographic models to tackle this question. The text is clear and concise, interpretations of the results are generally wise, and analyses well-conducted. I am convinced that this work will be a great addition to the speciation genomics literature, in particular because it highlights the unusual features of the marine realm and their consequences to speciation. I only have one main comment, and a few suggestions to improve the paper.
MAIN COMMENT
My main point is that the demographic analyses of post-divergence gene flow (L168-210), and its relation to species abundances (L212-287), could be improved.
I understand that the authors “only” have in hand a single genome per species to reconstruct their demographic history, and so this limits the type of demographic analyses that can be performed. In the paper, the authors employ the hPSMC method that uses a pseudo-diploid F1 hybrid genome to deduce the time of cessation of gene flow between species. This time is indirectly inferred based on its effect in reducing the estimate of divergence times. Then, to test for the influence of species abundance on hybridization, the authors estimate population sizes through time using PSMC, and discuss the two features (post-divergence gene-flow and species abundance).
First, I think that the connection between the two features could be made more readable. For example, you could add on Figure 3B,C,D the corresponding intervals for the cessation of gene-flow.
Second, as acknowledged by the authors their approach has some limitations, including that i) it does not provide the direction and rate of post-divergence gene-flow and, ii) it cannot disentangle a decline in species abundance vs. a reduction in interspecies gene flow. That is why I think it would be worth trying the Sequential Markovian Coalescent method implemented in MSMC-IM (Wang et al. 2020: doi:10.1371/journal.pgen.1008552). This will help to better evaluate the recent demography of each pair of species, including getting an estimate of both population sizes and migration rates over time. One difficulty is that this tool requires phased data, and so I am not sure whether the authors will have the relevant data for all species. Anyway, applying this tool to a single species pair for which phased data exist (or population data exists, so statistical phasing can be envisaged) would already be informative.
Third, for the specific question of the effect of species abundance on hybridization, another limitation that arises is the effect of confounding factors. Obviously, the number of barriers that accumulated in the genome of the species through time will also affect the probability of hybridization and/or introgression. How do you account for this effect? At least this point should be better discussed along lines L212-287.
MINOR COMMENTS
L285-287: the discrepancy between the hPSMC analysis (which shows that there is no ongoing introgression) and the presence of fertile contemporary hybrids is at odds. This could suggest that the hPSMC analysis is not appropriate to detect recent introgression events. Could you please further comment on that?
L448: Even if it seems obvious, it could help to indicate in the legend of Figure 2 that “divergence times” are in dark colours, and the “time interval during which gene flow ceased” is in light colour. Please, add an “s” to the time interval[s].
Supp. Tables S3, S4: I may miss something here, but I do not understand how the # of “Mapped reads” can be higher than the # of “Raw reads”?










