
Drift rather than host or parasite control can explain within-host Wolbachia growth
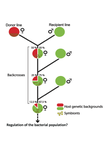
Wolbachia load variation in Drosophila is more likely caused by drift than by host genetic factors
Abstract
Recommendation: posted 04 May 2021, validated 11 May 2021
Duncan, A. and Hochberg, M. (2021) Drift rather than host or parasite control can explain within-host Wolbachia growth. Peer Community in Evolutionary Biology, 100126. https://doi.org/10.24072/pci.evolbiol.100126
Recommendation
Within-host parasite density is tightly linked to parasite fitness often determining both transmission success and virulence (parasite-induced harm to the host) (Alizon et al., 2009, Anderson & May, 1982). Parasite density may thus be controlled by selection balancing these conflicting pressures. Actual within-host density regulation may be under host or parasite control, or due to other environmental factors (Wale et al., 2019, Vale et al., 2011, Chrostek et al., 2013). Vertically transmitted parasites may also be more vulnerable to drift associated with bottlenecks between generations, which may also determine within-host population size (Mathe-Hubert et al., 2019, Mira & Moran, 2002).
Bénard et al. (2021) use 3 experiments to disentangle the role of drift versus host factors in the control of within-host Wolbachia growth in Drosophila melanogaster. They use the wMelPop Wolbachia strain in which virulence (fly longevity) and within-host growth correlate positively with copy number in the genomic region Octomom (Chrostek et al., 2013, Chrostek & Teixeira, 2015). Octomom copy number can be used as a marker for different genetic lineages within the wMelPop strain.
In a first experiment, they introgressed and backcrossed this Wolbachia strain into 6 different host genetic backgrounds and show striking differences in within-host symbiont densities which correlate positively with Octomom copy number. This is consistent with host genotype selecting different Wolbachia strains, but also with bottlenecks and drift between generations. To distinguish between these possibilities, they perform 2 further experiments.
A second experiment repeated experiment 1, but this time introgression was into 3 independent lines of the Bolivia and USA Drosophila populations; those that, respectively, exhibited the lowest and highest Wolbachia density and Octomom copy number. In this experiment, growth and Octomom copy number were measured across the 3 lines, for each population, after 1, 13 and 25 generations. Although there were little differences between replicates at generation 1, there were differences at generations 13 and 25 among the replicates of both the Bolivia and USA lines. These results are indicative of parasite control, or drift being responsible for within-host growth rather than host factors.
A third experiment tested whether Wolbachia density and copy number were under host or parasite control. This was done, again using the USA and Bolivia lines, but this time those from the first experiment, several generations following the initial introgression and backcrossing. The newly introgressed lines were again followed for 25 generations. At generation 1, Wolbachia phenotypes resembled those of the donor parasite population and not the recipient host population indicating a possible maternal effect, but a lack of host control over the parasite. Furthermore, Wolbachia densities and Octomom number differed among replicate lines through time for Bolivia populations and from the donor parasite lines for both populations. These differences among replicate lines that share both host and parasite origins suggest that drift and/or maternal effects are responsible for within-host Wolbachia density and Octomom number.
These findings indicate that drift appears to play a role in shaping Wolbachia evolution in this system. Nevertheless, completely ruling out the role of the host or parasite in controlling densities will require further study. The findings of Bénard and coworkers (2021) should stimulate future work on the contribution of drift to the evolution of vertically transmitted parasites.
References
Alizon S, Hurford A, Mideo N, Baalen MV (2009) Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. Journal of Evolutionary Biology, 22, 245–259. https://doi.org/10.1111/j.1420-9101.2008.01658.x
Anderson RM, May RM (1982) Coevolution of hosts and parasites. Parasitology, 85, 411–426. https://doi.org/10.1017/S0031182000055360
Bénard A, Henri H, Noûs C, Vavre F, Kremer N (2021) Wolbachia load variation in Drosophila is more likely caused by drift than by host genetic factors. bioRxiv, 2020.11.29.402545, ver. 4 recommended and peer-reviewed by Peer Community in Evolutionary Biology. https://doi.org/10.1101/2020.11.29.402545
Chrostek E, Marialva MSP, Esteves SS, Weinert LA, Martinez J, Jiggins FM, Teixeira L (2013) Wolbachia Variants Induce Differential Protection to Viruses in Drosophila melanogaster: A Phenotypic and Phylogenomic Analysis. PLOS Genetics, 9, e1003896. https://doi.org/10.1371/journal.pgen.1003896
Chrostek E, Teixeira L (2015) Mutualism Breakdown by Amplification of Wolbachia Genes. PLOS Biology, 13, e1002065. https://doi.org/10.1371/journal.pbio.1002065
Mathé‐Hubert H, Kaech H, Hertaeg C, Jaenike J, Vorburger C (2019) Nonrandom associations of maternally transmitted symbionts in insects: The roles of drift versus biased cotransmission and selection. Molecular Ecology, 28, 5330–5346. https://doi.org/10.1111/mec.15206
Mira A, Moran NA (2002) Estimating Population Size and Transmission Bottlenecks in Maternally Transmitted Endosymbiotic Bacteria. Microbial Ecology, 44, 137–143. https://doi.org/10.1007/s00248-002-0012-9
Vale PF, Wilson AJ, Best A, Boots M, Little TJ (2011) Epidemiological, Evolutionary, and Coevolutionary Implications of Context-Dependent Parasitism. The American Naturalist, 177, 510–521. https://doi.org/10.1086/659002
Wale N, Jones MJ, Sim DG, Read AF, King AA (2019) The contribution of host cell-directed vs. parasite-directed immunity to the disease and dynamics of malaria infections. Proceedings of the National Academy of Sciences, 116, 22386–22392. https://doi.org/10.1073/pnas.1908147116
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/2020.11.29.402545
Version of the preprint: https://www.biorxiv.org/content/10.1101/2020.11.29.402545v2
Author's Reply, 04 May 2021
Please find the new version of the manuscript.
We have made the suggested changes in the text and modified the title as the Reviewers suggested (highlighted in yellow in the attached tracked changes document).
We hope that this study will be suitable for recommendation by PCI.
Natacha Kremer
Decision by Michael Hochberg and Alison Duncan, posted 16 Apr 2021
Dear Authors,
The two reviewers were satisfied overall with your revisions and recommend that following these your preprint be approved by PCI. In parallel, Dr. Alison Duncan, who is co-recommending your preprint, has made numerous minor corrections to the manuscript that we hope you will consider in your revised submission. I will send the Word doc to you shortly.
Yours,
Michael
Download recommender's annotationsReviewed by Simon Fellous, 22 Mar 2021
The authors addressed my comments in a satisfactory fashion.
The only question that remains is whether this protocol enables disentangling drift from selection on Wolbachia density. Conspicuous selective factors, which would have varied among fly vials, could indeed explain the reported variations. I agree with the authors that abiotic factors, such as temperature, are unlikely candidates. However, I would imagine that among-vial variations of other biotic factors, such a bacterial gut symbionts, viruses other that DCV, or even transposable elements may have run unnoticed.
This comment should not prevent publication.
But, maybe would it be wise to change the title to something closer to the observations, for example, by highlighting the lack of effect of host genotype on Wolbachia density and octomom copy number.
https://doi.org/10.24072/pci.evolbiol.100368.rev21Reviewed by anonymous reviewer 1, 14 Apr 2021
I think the authors addressed all the comments raised on the previous version. Just one more comment: I find the title rather confusing. I suggest changing it to something explicit like “No evidence for host genotype effects on Wolbachia density” or “Wolbachia load variation in Drosophila is more likely caused by drift than host genetic factors”
https://doi.org/10.24072/pci.evolbiol.100368.rev22Evaluation round #1
DOI or URL of the preprint: 10.1101/2020.11.29.402545
Author's Reply, 17 Mar 2021
Decision by Michael Hochberg and Alison Duncan, posted 21 Jan 2021
Dear Dr. Kremer,
Your preprint has now been seen by two Reviewers. Their comments accompany this letter. Although both see interest in what your study attempts to achieve, they both have major reservations about the interpretation of the results. Both Reviewers question whether the study convincingly shows drift (and the absence of host genotype effects). Their arguments about possible alternative explanations for your results suggest that you will need to rethink your interpretations and revise accordingly, with the possible additional presentations/analyses suggested notably by Reviewer 2.
I would therefore encourage you to revise and resubmit your preprint to PCI Evolutionary Biology together with point-by-point replies to each of the reviewers’ comments. Please indicate in each reply where changes were made in the manuscript. Once your revision is received, we will contact both reviewers for their views on whether their concerns have been adequately addressed.
Yours sincerely, Michael Hochberg
Reviewed by anonymous reviewer 1, 24 Dec 2020
In this study, the authors introgressed a Wolbachia strain into six different (previously uninfected) Drosophila lines to evaluate if host genotype has an influence on Wolbachia loads and Octomom copy numbers within Wolbachia genomes (octomom copy numbers were previously shown to be positively correlated with Wolbachia loads). Although Wolbachia loads significantly varied among introgression lines, the authors could not conclude that host genotype affected Wolbachia loads because only a single introgression line per host genotype was generated. The authors therefore generated three replicate introgression lines for two host genotypes in a second experiment, as well as reciprocal crosses between the two host genotypes. Because they found extensive load variation among replicate lines withing genotypes, as well as variation over time within lines, they conclude that host genotype has no effect on Wolbachia load. Instead, they suggest that load variation is a consequence of drift.
Although I am generally strongly in favor of publishing “negative results”, I am not convinced that the study shows absence of host genotype effects on Wolbachia loads. The data also do not show that load variation is caused largely by drift (although I agree with the authors that the initial endosymbiont load in eggs may indeed be quite important, the data do not show this). I do agree with the authors that the variation among generations and replicate lines within the two genotypes is striking. But the authors only analyze two host genotypes with multiple replicates for introgression crosses, and based on a sample of n=2 I would not conclude absence of host genetic background effects. Furthermore, we do not actually know the nuclear genome composition of the introgression lines (Fig. 1 is an expectation for neutral polymorphisms…). For example, there could be selection for the retention of w1118-derived haplotypes during introgression of Wolbachia, or presence/absence of w1118-derived haplotypes in females could actually influence Wolbachia loads. Thus, different replicate lines within genotypes could fix w1118 alleles in different regions following the arrest of introgression. I don't know how likely this is, but hope these examples illustrate the different reasons why I am not convinced this study shows that host genotype is not an important driver of endosymbiont load.
It would also be helpful if the authors could clarify sample sizes. For example, how large were the population sizes during introgression and during the maintenance of the introgression lines? What was the protocol for the maintenance (eg. would it be possible than only a small portion of females contributed offspring to the next generation?). This information would be useful to assess the potential for drift. Also, why waiting 8 generations after the introgresson protocol to measure Wolbachia load? How many individuals per strain were used to quanitify Wolbachia loads and octomom copies? etc.
Minor comments.
I am not sure what to make from the positive correlation between load and octomom copy number. What does it mean exactly? Do the authors find anything new here with respect to previous studies? Or is it an independent confirmation of the same pattern (also useful) – would be helpful if this could be clarified.
I suggest revising the statement L240 “…These results suggest that different host genetic backgrounds selected specific variants of the symbiotic community. However, other factors, like genetic drift through a founder effect during the vertical transmission of symbionts from the donor line and / or from one host generation to the other, could also explain this pattern.” The results are “consistent with”, but since the authors later revise their conclusion, better not first suggest the results are caused by host genetic effects.
L11 density of endosymbiont density
L17 I would say “between host individuals”
Some explanation for the positive correlation of octomom copies and bacterial density as a consequence of drift would be useful in the abstract
L82 flies “with endosymbionts” harboring more copies of Octomom (note that throughout the text, it is often not clear, from the formulation, whether the flies or Wolbachia have octomom copies).
L89 symbionts (plural)
Figure 1, second panel doesn’t really depict a reciprocal cross (but a unidirectional introgression cross as in the first panel).
L130 explained (past)
L140 and following (Quantification of wMelPop density and Octomom copy number): I suppose this was done on female flies only? (please clarify)
L211-212 reformulate to “differ significantly among introgression lines” – the statement “are significantly influenced by the host genotype” goes against the author’s own interpretations and would in fact require replicate introgression experiments per genotype.
L230 remove double citation of the same paper
Reviewed by Simon Fellous, 20 Jan 2021
In this manuscript Bénard et al. describe the results of a series of experiments exploring the genetic determinants of intra-cellular symbiont regulation in insect hosts. Using the Drosophila melanogaster-Wolbachia system, the authors tested whether host genotype controls symbiont cell density. This work builds upon previous studies from these and other authors that established a link between the number of copies of the Octomom gene in Wolbachia and symbiont density. The present study did not observe stable host control of symbiont numbers over time, suggesting drift largely determines how many symbionts may be found in hosts. The experiments and analysis appear sound and the writing is clear. It is a well thought study. I only have comments on the discussion/interpretation of the data and would welcome a couple of additional analyses and figures. Once these points are addressed, the manuscript should make an excellent contribution to PCI Evol Bio and the field in general. Major comments: . The conclusion that drift during maternal transmission is the main factor at play is based on the lack of effect of host genetics on Wolbachia density. However, you did not discuss the possibility that unidentified selective pressures drove these variations. External selective factors could indeed vary over time and among fly lines/fly vials. For example, Rohrscheib et al. (2016) argue an abiotic factor such as temperature can drive Wolbachia density variations. Similarly, these bacteria being involved in anti-viral defences, outbreaks of viruses in fly vials could have run unnoticed. Incidentally, it would explain why symbiont densities reached peaks and dropped simultaneously in some line replicates. Do you have reasons to exclude selection by external factors as a determinant of Wolbachia density? . Along these lines, do you exclude the possibility that a gene or set of Wolbachia genes different from Octomom determined symbiont density? In that case maybe did these factors evolved quickly. . The initial relationship between Octomom copy number and Wolbachia density is useful to formulate your hypotheses (Fig 3). It would be equally useful to present this correlation, or lack of, in the final sets of data (from Fig 4 and 5). Btw, do you have individual data? This would give an idea of within-host line variation? Does the correlation stand at the scale of individuals as well as populations? . L 401-404: association between mean symbiont numbers and variance is reported but not statistically tested nor illustrated. Can you please provide support to this element of discussion? Also, does the relationship stand when data is log-transformed? … simply because greater variance is to be expected when mean increases. . L 410-414: is it possible to distinguish transmission drift from host population drift? One is necessarily nested into the other. A naïve expectation (my naïve expectation to be fair) would be that even if transmission drift was high, a lack of pop drift should maintain mean symbiont density due to a population scale averaging effect. In other words, transmission drift may only drive trait variation if associated with subsequent host population bottlenecks, that is with population drift. Minor suggestions : L 19 : “between partners” please be more specific L 42-43: this description better matches horizontally transmitted parasites that vertically transmitted ones. Maybe talk about reproductive output? Baby numbers... L 44: rather that density, maybe use reproduction rate or resource exploitation rate. These terms are compatible with macroparasites which numbers don’t change even though they extract more or less resources from their host. L96: please give equivalent location precision for all pops Simon Fellous
https://doi.org/10.24072/pci.evolbiol.100368.rev12









