
Macroevolutionary drivers of brain evolution in primates
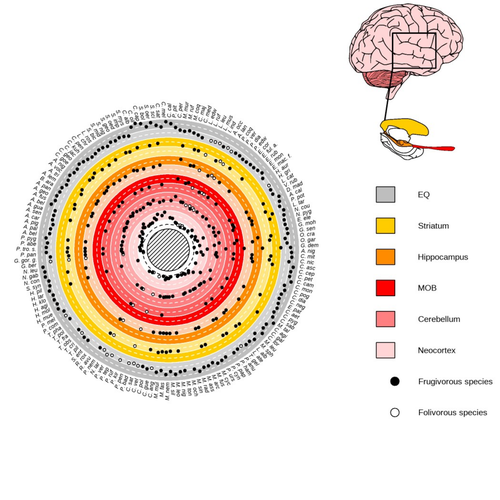
Primate sympatry shapes the evolution of their brain architecture
Abstract
Recommendation: posted 25 February 2023, validated 28 February 2023
Condamine, F. (2023) Macroevolutionary drivers of brain evolution in primates. Peer Community in Evolutionary Biology, 100548. https://doi.org/10.24072/pci.evolbiol.100548
Recommendation
Studying the evolution of animal cognition is challenging because many environmental and species-related factors can be intertwined, which is further complicated when looking at deep-time evolution. Previous knowledge has emphasized the role of intraspecific interactions in affecting the socio-ecological environment shaping cognition. However, much less is known about such an effect at the interspecific level. Yet, the coexistence of different species in the same geographic area at a given time (sympatry) can impact the evolutionary history of species through character displacement due to biotic interactions. Trait evolution has been observed and tested with morphological external traits but more rarely with brain evolution. Compared to most species’ traits, brain evolution is even more delicate to assess since specific brain regions can be involved in different functions, may they be individual-based and social-based information processing.
In a very original and thoroughly executed study, Robira & Perez-Lamarque (2023) addressed the question: How does the co-occurrence of congeneric species shape brain evolution and influence species diversification? By considering brain size as a proxy for cognition, they evaluated whether species sympatry impacted the evolution of cognition in frugivorous primates. Fruit resources are hard to find, not continuous through time, heterogeneously distributed across space, but can be predictable. Hence, cognition considerably shapes the foraging strategy and competition for food access can be fierce. Over long timescales, it remains unclear whether brain size and the pace of species diversification are linked in the context of sympatry, and if so how. Recent studies have found that larger brain sizes can be associated with higher diversification rates in birds (Sayol et al. 2019). Similarly, Robira & Perez-Lamarque (2023) thus wondered if the evolution of brain size in primates impacted their dynamic of species diversification, which has been suggested (Melchionna et al. 2020) but not tested.
Prior to anything, Robira & Perez-Lamarque (2023) had to retrace the evolutionary history of sympatry between frugivorous primate lineages through time using the primate tree of life, species’ extant distribution, and process-based models to estimate ancestral range evolution. To infer the effect of species sympatry on the evolution of cognition in frugivorous primates, the authors evaluated the support for phylogenetic models of brain size evolution accounting or not for species sympatry and investigated the directionality of the selection induced by sympatry on brain size evolution. Finally, to better understand the impact of cognition and interactions between primates on their evolutionary success, they tested for correlations between brain size or species’ sympatry and species diversification.
Robira & Perez-Lamarque (2023) found that the evolution of the whole brain or brain regions used in immediate information processing was best fitted with models not considering sympatry. By contrast, models considering species sympatry best predicted the evolution of brain regions related to long-term memory of interactions with the socio-ecological environment, with a decrease in their size along with stronger sympatry. Specifically, they found that sympatry was associated with a decrease in the relative size of the hippocampus and striatum, but had no significant effect on the neocortex, cerebellum, or overall brain size.
The hippocampus is a brain region that plays a crucial role in processing and memorizing spatiotemporal information, which is relevant for frugivorous primates in their foraging behavior. The study suggests that competition between sympatric species for limited food resources may lead to a more complex and unpredictable food distribution, which may in turn render cognitive foraging not advantageous and result in a selection for smaller brain regions involved in foraging. Niche partitioning and dietary specialization in sympatry may also impact cognitive abilities, with more specialized diets requiring lower cognitive abilities and smaller brain region sizes.
On the other hand, the absence of an effect of sympatry on brain regions involved in immediate sensory information processing, such as the cerebellum and neocortex, suggests that foragers do not exploit cues left out by sympatric heterospecific species, or they may discard environmental cues in favor of social cues.
This is a remarkable study that highlights the importance of considering the impact of ecological factors, such as sympatry, on the evolution of specific brain regions involved in cognitive processes, and the potential trade-offs in brain region size due to niche partitioning and dietary specialization in sympatry. Further research is needed to explore the mechanisms behind these effects and to test for the possible role of social cues in the evolution of brain regions. This study provides insights into the selective pressures that shape brain evolution in primates.
References
Melchionna M, Mondanaro A, Serio C, Castiglione S, Di Febbraro M, Rook L, Diniz-Filho JAF, Manzi G, Profico A, Sansalone G, Raia P (2020) Macroevolutionary trends of brain mass in Primates. Biological Journal of the Linnean Society, 129, 14–25. https://doi.org/10.1093/biolinnean/blz161
Robira B, Perez-Lamarque B (2023) Primate sympatry shapes the evolution of their brain architecture. bioRxiv, 2022.05.09.490912, ver. 4 peer-reviewed and recommended by Peer Community in Evolutionary Biology. https://doi.org/10.1101/2022.05.09.490912
Sayol F, Lapiedra O, Ducatez S, Sol D (2019) Larger brains spur species diversification in birds. Evolution, 73, 2085–2093. https://doi.org/10.1111/evo.13811
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Both authors were supported by a doctoral grant from the École Normale Supérieure, Paris.
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/2022.05.09.490912
Version of the preprint: 2
Author's Reply, 14 Feb 2023
Decision by Fabien Condamine, posted 28 Nov 2022, validated 28 Nov 2022
Dear Benjamin and Benoît,
Thank you for submitting your revised work (manuscript #548) to PCi Evol. Biol.
I have sent back for re-review to the previous 5 expert reviewers. We have now received the comments from 4 of them, which you will find associated to this decision.
All the reviewers expressed again positive comments about the manuscript, on which I agree, and 3 referees added a few more comments that should not be the most difficult to address.
However, I also agree with the remaining comments raised during this new round of review, and I would like to see a revised version of the manuscript. I think it's important to clarify several aspects of the study as suggested by the reviewers.
After this revision, I am opitmistic that it will lead to a recommendation since I think I will not send it back for re-review (the referees have already done a great job, and I thank them for it). I hope these reviews will help you to improve your study.
I look forward to see a revised version of your manuscript. If you have any questions, please do not hesitate to reach me.
All the best,
Fabien Condamine, for PCi Evol. Biol.
Reviewed by anonymous reviewer 1, 26 Nov 2022
I have read with great interest the revised version of the manuscript Primate sympatry shapes the evolution of their brain architecture. I thank the authors for their positive response to previous comments and commend them for the hard work addressing them. I think the work is clearer, however I still have a few questions that I believe need to be addressed before the work is suitable for recommendation.
Firstly, to me it is not clear why competition is expected to lead to directional changes, i.e. either enlarged or decreased brain structure or brain size. As I understand it competition may lead to character displacement, to avoid competing for specific resources with members of a given species. However, not all competing individuals are predicted to modify specific characters and even less so in the same direction and degree. Indeed, individuals of the more successful competitor would face no selection to change. Thus, I wonder if the prediction resulting from high sympatry might not be increased variance in brain structures or brain sizes across a set of sympatric species, rather than enlarged or reduced brain structure or brain sizes. In support of this idea, if I am not mistaken, this is what the authors actually found for body size, a trait that is probably more directly linked to resource use in primates (or at least the Discussion seems to suggest this in lines 497-500).
The point above leads me to my second point of concern. The authors have chosen to use both body size and whole brain size to control for allometric effects in their analyses of brain structure sizes. However, I was left wondering whether this is justified given they found that competition actually impacts body size evolution, as would be predicted, but actually rather differently than they predict for brain structure sizes. I acknowledge that the fact that results are the same when using either variable as a control for allometric effects, which I honestly found a bit puzzling given the effect of competition on body size evolution.
I also found somewhat surprising that the authors propose that competition could lead to reduced brain structure or brain sizes due to depleted resources and the energy constraints of maintaining a large brain. There is indeed plenty of evidence supporting the claim of high costs of brain growth, development and maintenance. However, competition would result in depleted resources for individuals that are less successful in competing but not necessarily for all competitors. This would also depend on the degree to which resources can be defended and thus how competition plays out. In fact, whether resources are defendable or not would actually have a very strong effect on the predicted outcome of sympatry and thus resource competition. There is no mention regarding whether resources can be defended or not. I.e. can members of one group effectively monopolize one or a group of fruiting trees impeding individuals of other species to access the resource? Or rather, is competition more scramble where arriving first gives precedence and thus greater access to resources and groups leave to search for other food sources when too many groups arrive, such as in a producer scrounger game?
In the Discussion the authors state that their results clearly emphasize the compromise due to high energetic demands of the brain as the largest structures, the cerebellum and neocortex, as well as whole brain size, are best fitted by an OU model. They propose that this may suggest a stabilization towards an optimal size resulting from an equilibrium between costs and benefits. It is not clear to me why they expect the optimum to be the same for all species. Certainly the benefits of a given relative brain size will differ among species given differing selection factors. Costs could also potentially differ if species are not all equally successful in acquiring resources. Finally, great care must be taken when interpreting the results of these models, one thing is the pattern that is observed but another is the process that may be responsible for said pattern (see e.g. Revell et al 2008). Furthermore, there are studies suggesting care must be taken when interpreting the results of OU models (see e.g. Cooper et al. 2015).
Reviewed by Paula Gonzalez, 12 Nov 2022
Reviewed by anonymous reviewer 2, 07 Nov 2022
This is the revised version of the previous manuscript. The first version was already very good, but I had a few concerns mainly regarding simpatry and niche partitioning. I think in this new version these concerns were satisfactorily tackled when possible or at least acknowledged when the problem was not immediately solvabl . I really like the final result and I think this will be a valuable contribution to the literature. While some doubts might remain regarding how convincing the results are, I think it is now up to the readers to make that call. The authors did a good work presenting the limitations and a balanced and honest interepretation. Also, I also was satisfied reading the author's response to the other reviewer's concerns, which I shared. So I have no further comments and I would definitively reccomend the publication of such a valuable contribution to macroevolutionary studies on primate brain evolution.
Thanks for inviting me to review this interesting manuscript.
https://doi.org/10.24072/pci.evolbiol.100548.rev23
Reviewed by Orlin Todorov, 28 Nov 2022
Hello,
here is my second round of review of the manuscript "Primate sympatry shapes the evolution of their brain architecture" submitted to PCI Evol Bio. Detailed comments can be found in the attached pdf file.
Cheers,
Dr Orlin S. Todorov
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2022.05.09.490912
Version of the preprint: 1
Author's Reply, 03 Oct 2022
Decision by Fabien Condamine, posted 04 Jul 2022
Dear authors,
Thank you for submitting your work (manuscript #548) to PCi Evol. Biol., which is much appreciated.
We have now received the comments from 5 expert reviewers (Dr. Orlin Todorov and four anonymous reviewers), which you will find associated to this decision. All five reviewers expressed positive comments about the manuscript, on which I agree.
However, I also agree with the comments raised during this round of review, and I would like to see a revised version of the manuscript. I think it's important to have clear definition of sympatry/co-existence to make predictions clearer. The definition of diet (frugivory) can be important to clarify too, especially to make improvements comapred to previous studies. In addition, there is apparently some issues with data combination, and a few issues with the analyses of the data. I do think these comments are crucial to take into account. The recent primate tree of Wisniewski et al. (2022 - PRSB) has been proposed, but I have important reservations about this tree. I reviewed this paper and many problems remains in the tree. Also their biogeographic analyses are poor, but I agree that this should be discussed in light of your inferences. I also think a revised title would better reflect the results.
Overall, I am opitmistic that such a revision process can lead to a recommendation but the study first needs improvements to attain this point. I hope these reviews will help you to improve your study.
On a personal opinion, I really enjoy the study and I concour with the general opinion that the manuscript addresses a passionating question, is already good and of high quality. I am impressed to see the amount and quality of data put together to address the question. As a macroevolutionary biologist, I remain a bit disappointed with the lack of figure showing the diversification rates over time. I see you have one in the Supplementary Material, what about using this to draft a figure for the main text? I know you have tables reporting the results for the diversification rates. I would also recommend the authors to put the ClaDS tree in the Supplementary Material, or along the figure with rates through time in a min-text figure. A last point that would be worth looking at is the fossil record of primates. I leave this opportunity open to the authors but in macroevolution, fossils provide direct evidence for presence and species' traits.
I look forward to see a revised version of your manuscript. If you have any questions, please do not hesitate to reach me.
All the best,
Fabien Condamine, for PCi Evol. Biol.
Reviewed by anonymous reviewer 3, 23 Jun 2022
Comments to the Author
Review of “Species sympatry shapes brain size evolution in Primates” (PCIEvolBiol #548)
This is an interesting manuscript that explores an important and controversial topic in evolutionary neurobiology – the selective forces shaping brain size and structure variation in nonhuman primates. The goal of this manuscript is to test whether sympatry predicts species differences in brain size or the size of certain brain regions (in frugivorous primates specifically). To accomplish this, they assemble new neuroanatomical data and apply phylogenetic comparative methods. They conclude that sympatry leads to a reduction in the size of the hippocampus and striatum, and that species in sympatry diversified more slowly.
This manuscript is very well written, and the analyses are thoughtfully executed. However, there some issues with the methods used (e.g., use EQ as a dependent variable, pooling of data from methodologically distinct resources, choice of brain regions to analyze). Accordingly, I suggest major revisions to this manuscript – my feedback it organized by section below.
Title
- I believe the title should more accurately reflect the results. In particular, the results suggest sympatry may affect the relative size of specific brain regions, not overall brain size, so a title of “Species sympatry shapes brain structure evolution in Primates” (or something similar) would be more appropriate.
Introduction
- The authors provide extremely clear and well-structured hypotheses and predictions. This is pretty rare in comparative work and is very much appreciated by this Reviewer.
- Prediction 2: Given that this prediction is centered on sensory information processing, why not examine V1 and AOB (instead of the whole neocortex)? Data is available for these areas.
- Prediction 3: What about the role of the AOB in social contexts?
Methods
- The methods are clearly explained.
- The Navarette data should only be combined with other datasets for the neocortex and cerebellum, but not for the hippocampus or striatum (see their Erratum: https://doi.org/10.1159/000496658).
- Do % of overlapped range and # of sympatric species covary? It would be informative to see the results with these predictors modelled separately.
- PGLS models:
- EQ should not be used as a dependent variable to model relative brain size. Rather, body size should be included as a covariate and brain size as a dependent variable.
- It may be more appropriate to model region size relative to rest of brain size (rather than body size), since if sympatry drives selection on body size, this could confound results.
Results
- The results are clear and well-written.
- It is not accurate to report significant results for the hippocampus when p = 0.058 (>0.05), unless the authors are using a different significance cutoff.
Discussion
- Line 383: “Bigger brains are not necessarily better” – when comparing brains across species, we also need to consider species differences in brain composition
- Line 409: “As a result, it might even generate a selection for smaller brains.” – There is no evidence for this.
- How do the reported results for the hippocampus relate to previous work on the socioecological correlates of this region (Todorov et al. 2019; DeCasien et al. 2019; Schilder et al. 2019)?
I hope the authors find these comments helpful in revising their manuscript.
https://doi.org/10.24072/pci.evolbiol.100548.rev11
Reviewed by anonymous reviewer 2, 13 Jun 2022
This a very interesting manuscript where the authors test the hypothesis that species competition for resources in Primates differentially impacts the evolutionary trajectories of the (relative) sizes of several brain regions. To do this they compile a comparative dataset of trait (brain regions’ sizes), distribution, and phylogenetic data, and implement a battery of phylogenetic comparative methods mostly based on evolutionary modelling. The paper is carefully and well written, it is methodologically very sound and it is a fantastic example of a well conducted study based on a phylogenetic comparative method approach. I highlight the careful consideration the authors had for dealing with uncertainty both in the data as in the analytical pipeline, which is rare to see in the field.
Nonetheless, I have a main concern regarding the author’s approach. While I admit that the hypothesis is very intringuing, I suspect that there could be a significant conflation of sympatry with competition. First of all, because current sympatry might reflect coexistence, not competition. Second, and specially in Primates, under the label of “frugivory” there is a plethora of different dietary strategies, as basically all Primates could be considered frugivorous animals. Although this is a problem that many other papers also have, here it could be potentially more problematic as what the authors are trying to model explictly are the effects of competition on cognition. But if there’s no or little resource compsumption overlap, it could be argued that there would be hardly any competition despite sympatry. In fact, it is known that currently sympatric primates tend to partition niche space quite considerably. By looking at the data it is not completely clear for me which species were finally considered as frugivorous, but for example, all or most platyrrhine primates seem to be above the 20% threshold for being considered frugivorous. However, this hides the fact that some species specialize in eating insects and tree exudates (Callithricins), others are seed predators (Pithecids, they forage for hard fruits impossible to eat for other species to get to the seeds through morphological adaptations) and others (Alouatta) rely heavily and seasonally on leaves without being as “folivorous” as colobines. Moreover, they segregate vertically in the three-dimensional canopy. This is not to say that there’s no competition or that there wasn’t in the past, and while I don’t have a clear solution to this problem (and I aknowledge that the author's used two different thresholds for assignign categories), I would like to see a more nuanced discussion and interpretation of results regarding this possibility. Big dietary categories are useful for macroecological studies as they allow to incorporate a lot of data, but the lack of detail perhaps is at some point harmful. I would like to see the authors thoughts on this reflected in the paper. Nonetheless, that the niche partitioning in primates could have also a “cognitive dimension” is a possibility that has been rised previously in other primate brain studies too (see for example Aristide et al 2016 PNAS for brain evolution in platyrrhine primates).
Additionally, and although I think the authors did their best to tackle uncertainty regarding biogeographic reconstructions, explictly akwnoledging the limits of extant-only/neontological approaches would be recommended. For example, the authors would not want to miss the recent paper by Wiesniecki et al (2022) PROC. B: https://royalsocietypublishing.org/doi/full/10.1098/rspb.2021.2535
https://doi.org/10.24072/pci.evolbiol.100548.rev12Reviewed by anonymous reviewer 1, 13 Jun 2022
I have read with interest the preprint "Species sympatry shapes brain size evolution in Primates". I have several comments and suggestions which I hope the authors will find useful to improve their work.
Major comments:
My first concern regards the hypothesis, as it is unclear why sympatry is expected to influence brain evolution. As noted by the authors, the social brain hypothesis has been porposed as a general explanation for brain evolution. I note that although the hypothesis was initially developped with primates in mind, it has been extended to other vertebrates (see e.g. Dunbar and Shultz 2007), although support for it beyond primates is mixed. The social brain hypothesis suggests that more complex interactions among members of a given species will lead to selection for greater cognitive abilities. The emphasis of the social brain hypothesis, as I understand it at least, is on the interactions among members of a given group. In fact, part of the reason of the mixed support in non-primate vertebrates is the fact that group size per se is actually a rather bad proxy for social complexity, and we lack the detailed observations of social interactions among group members that exist for primates in other species. The social brain hypothesis might well not apply to non-primates, but some of the tests likely do not capture its essence, that it that complex social interactions favor larger brain size. In fact, in their review Dunbar and Shultz (2007) propose different proxies for social complexity for non-primates (see e.g. Fig 3). Thus, following from this argumentation, sympatry is just as likely a bad proxy for interspecific interactions between sympatric species, as some sympatric species are unlikely to interact much while others might do so much more. This is one of the reasons why I find the argumentation presented in the manuscript unconvincing.
Secondly, and related to the point above, the authors propose that sympatry could both lead to increased or decreased sizes of brain structures, which makes me wonder how they expect to actually reject their hypothesis, if any relationship between sympatry and brain structure size would support it. The argument in favour of decreased brain size is actually rather weak, as they suggest high competition would lead to reduced resource availability and thus less investment into brain size. This seems somewhat far-fetched, as studies have proposed a diversity of means by which individuals may mitigate the negative effects of interspecific competition for resources, through behavioural changes, or changes in which resources are exploited or prefered. A reduction in brain size seems like quite a high price to pay, especially given the rather well documented benefits of enlarged brain size (see e.g. Benson-Amram et al. 206, Sol et al. 2005, Sol et al. 2008, Jimenez-Ortega et al. 2020, to name a few). In addition, and perhaps more importantly, it is unclear why sympatry would lead to changes in brain size at the species-level, rather than increased variance among populations, based on the degree of / number of sympatric species. What I mean is that if sympatry causes competition, it would be only the populations that compete with other species for food which would be under the selection pressure to modify brain size / brain structure size, it is unclear why populations not under said selection pressure would change at all. For example, populations of two sympatric species show greater pre-cigotic isolation than allopatric populations of the same species. In addition, body size is not taken into account in the hypothesis, but I would imagine that larger species are likely to be much less affected by sympatry than smaller ones, as the larger species will be much more efficient at defending a limited resource against heterospecific competitors than smaller species, hence the effect of sympatry might not be equal across all species.
I also have important concerns regarding the data and methods used to test the hypothesis.
Firstly, brain size, and more importantly brain structure sizes, were collated from different sources. How did you ensure that the data were comparable? Methods, sample sizes, etc likely differed across studies. How did you deal with sexual size dimorphism? Were data collated for both males and females? I imagine all were adult specimens?
For brain size the authors estimate encephalization quotients, but use of an ANCOVA approach would be much more appropriate from a statistical point of view (see e.g. Freckleton 2002, García-Berthou 2001, Nakagawa et al. 2017).
For brain structure sizes the authors use a ratio between brain volume and body mass, which is much more worrying given that relative brain size is assumed to vary among species, as they calculate EQ. If relative brain size is assumed to vary among species, and the authors assume that there is at least some degree of mosaic evolution of brain structures, whole brain size would be the appropriate means to adjust structure sizes based on allometry, i.e. the expectation is that the structure size will respond to changes in brain size or at least will change in size relative to the whole brain. Again, as mentioned above, analyses should be done including brain size as a co-variate in all analyses.
To estimate sympatry the authors used distribution maps from the IUCN red list. However, the maps are estimates of species distributions, but not always specifically based on actual observations of species. How did the authors verify that said maps coincided with actual registered sightings of the species, especially given these were used to estimate sympatry.
I also worry about the use of biogeographic analyses to estimate 'history of sympatry' along the primate phylogeny. Such reconstructions of ancestral distributions make the assumption that the factors that determine a species' distribution range have remained stable from present to the past, which seems unlikely.
In the Methods section the authors surprise readers by anouncing that they also checked for correlative patterns between primate brain size an ddiversification rates, however this does not relate to their original hypotheses, or it is unclear how it relates to them. I would suggest that either they include this question in the Introduction or just leave it out, as it seems rather tangential to the hypothesis being tested.
In line 208-209 the authors state that they "fitted phylogenetic models the evolution of the size of different brain areas independently". However, this assumes completely moisaic evolution of brain structure sizes, which is not the case for primates (see e.g. Barton and Harvey 2000, Finlay and Darlington 1995). Furthermore, some of the models of trait evolution are specifically designed to test whether inter-specific competition could influence phenotypic evolution, however as I argue above, I would expect changes in particular functional traits linked to resource acquisition, it is unclear why brain size, or brain structures, would respond directly to competition. The DD models assume rates of phenotypic evolution are influenced by species richness (see Morlon et al 2016), but these are based on theory on adaptive radiations, where available niches become filled as the radiation progresses (and lineage diversity increases) and thus niche partition becomes increasingly finer, all of which is expected to influence the rate of phenotypic evolution. It is not clear how these models relate to the hypothesis being tested here.
Finally, in the Results the authors first find that models that do not include sympatry best explained the evolutionary history of EQ, neocortex and cerebellum. They apparently also fitted OU models to explain trait evolution, however it is unclear why they expected an OU model (especially with a single optimum value) to explain evolution of brain and brain structure sizes. In addition interpretation and fit of such simple OU models has been questioned (see Cooper et al. 2015).
https://doi.org/10.24072/pci.evolbiol.100548.rev13
Reviewed by Paula Gonzalez, 19 Jun 2022
The paper aims at evaluating the effect of sympatry on the evolution of brain size in Primates species. The main assumption of the study is that cognitive abilities are associated with the number of species sharing the same environment due to competition for food between species and direct and indirect interactions between heterospecifics. The topic of brain evolution in Primates has been extensively studied, and the role of ecological and social factors has been tested with variable results according to the sampling design. In this context, the topic of this study is important because it provides a new perspective focusing on the interaction among primate species. However, I found that some ideas and criteria used to define the variables under study should be better explained (see comments below).
In the Introduction, the authors should clearly explain the definition of sympatry used throughout the study. According to the methods used, it seems sympatry is defined as species sharing the same biogeographic area. Given the wide geographic extension of the areas analyzed, the inclusion of two species as sympatric not necessarily implies a direct interaction between them (e.g., Ayres and Clutton-Brock, 1992). Consequently, the hypotheses proposed, which assume a competition for food and direct and indirect interactions between heterospecific sympatric species, are hard to test based solely on such biogeographic areas. At least, the caveats of this approach need to be discussed.
The hypotheses tested assume a direct association of each brain area with only one function, which is problematic because some brain areas include several regions with different functions. For example, hypothesis #2 assumes that the neocortex is associated with processing immediate sensory information (such as the primary motor and visual cortex) even though it also comprises multimodal areas. Similarly, the hippocampus is not only related to spatial navigation and memory but is organized into regions with different functional specializations (see Todorov et al., 2019).
Regarding the material and methods, the study relies on publicly available data on brain size and body mass to estimate the EQ and the relative size of five brain regions. The authors estimated the relative size of the different brain areas relative to body size instead of brain size. There are different approaches to estimating relative brain size and there are no a priori criteria to prefer one instead of the other but depends on the aim of the study. In this sense, brain size relative to body mass is usually used to discuss the resource allocation between tissues while the second approach is used in the framework of mosaic brain evolution. Given the hypotheses tested, the latter method seems to be more informative, although the authors might have good reasons to prefer the other method, which needs to be better explained. Moreover, brain size relative to body mass can vary due to changes in body size (Smaers et al., 2021), although the latter variable was not included in the models. Particularly in platyrrhines, previous studies show that the evolution in body size was the main factor responsible for changes in phenotypic traits, including the skull, a structure closely related to the brain (Marroig and Cheverud, 2001). The allometric effect of body size should be taken into account along with other variables such as group size, which has also been related to brain evolution in primates (Dunbar, 1998; among others). The probable confounding factor of group size for testing Hypothesis #3 should be at least discussed.
Minor comments. Figure 3 is difficult to read, and the relative brain size among species is hard to compare.
References:
Ayres, J. M., & Clutton-Brock, T. H. (1992). River boundaries and species range size in Amazonian primates. The American naturalist, 140(3), 531–537. https://doi.org/10.1086/285427
DeCasien, A., Williams, S. & Higham, J. (2017). Primate brain size is predicted by diet but not sociality. Nat Ecol Evol 1, 0112. https://doi.org/10.1038/s41559-017-0112
Dunbar, R. I. M. (1998) The social brain hypothesis. Evol. Anthropol. 6, 178–190.
Marroig, G., & Cheverud, J. M. (2001). A comparison of phenotypic variation and covariation patterns and the role of phylogeny, ecology, and ontogeny during cranial evolution of new world monkeys. Evolution; international journal of organic evolution, 55(12), 2576–2600. https://doi.org/10.1111/j.0014-3820.2001.tb00770.x
Smaers, J. B., Rothman, R. S., Hudson, D. R., Balanoff, A. M., Beatty, B., Dechmann, D. K. N., … Safi, K.(2021). The evolution of mammalian brain size. Science Advances, 7(18), 9. https://doi.org/10.1126/SCIADV.ABE2101/SUPPL_FILE/ABE2101_SM.PDF
Todorov, O. S., Weisbecker, V., Gilissen, E., Zilles, K., & de Sousa, A. A. (2019). Primate hippocampus size and organization are predicted by sociality but not diet. Proceedings of the Royal Society B, 286(1914). https://doi.org/10.1098/RSPB.2019.1712
https://doi.org/10.24072/pci.evolbiol.100548.rev14
Reviewed by Orlin Todorov, 07 Jun 2022
Thanks for inviting me to review the manuscript entitled "Species sympatry shapes brain size evolution in Primates". The paper is well written, the problem is tackled appropriately through the application of state-of-the-art phylogenetic comparative methods and the sample collated from the literature is comprehensive. The chosen approach, namely to look into brain partitions besides whole (relative) brain size is commendable. I am providing line-to-line comments here and the lines referred to have been highlighted in the pdf file I am attaching.
I consider the paper to be worthy of publication in a high-ranking journal, but there are a few problems that need to be addressed. Some of the reasoning in the introduction might need to be addressed (i.e. sympatry or congener sympatry?), some models might need to be reanalysed (see my comments regarding normalisation and derivation of EQ) and there might be methodological solution already available that directly address the problem with sample size (as the authors sacrifice the majority of their sample size in most of their analyses). I also have reservations regarding the use of ancestral state estimation without calibrating with data from the fossil record (in this case 'diet').
Comments:
Title: Primates does not need capitalisation.
Ln12 – For an opening statement, this is not necessarily true. Some of ‘the main hypotheses’ emphasize the role of the environment.
Ln42 – This seems to be as valid not only for other primate species, but for many other animals, i.e. birds, other mammals etc that will also compete with primates for similar food sources. How can/would one account for that? If the problem is phrased as ‘sympatry’ then most definitely competition between primates and other frugivorous species must be taken into account? Maybe rephrase the whole issue as ‘congeners sympatry’ also in the title?
Ln56 – This is a really good point! I like the idea of looking into different brain partitions instead of whole brain / relative brain size.
Ln62 - I don’t think you need to capitalize brain partitions.
Ln71 – But also the hippocampus? It has been shown that hippocampal volume is related to sociality in primates (Todorov, 2019 – it is in the citation list of the manuscript).
Ln81 – In terms of memory and primates, using a proxy (as hippocampus) might be ok (can you refer to a paper in the manuscript showing that there is a correlation between memory performance and hippocmpal size in primates?), but we do have direct measures of memory for many primates – MacLean, 2014 (MacLean, E. L., Hare, B., Nunn, C. L., Addessi, E., Amici, F., Anderson, R. C., ... & Zhao, Y. (2014). The evolution of self-control. Proceedings of the National Academy of Sciences, 111(20), E2140-E2148.)
Ln100 – Just humans should be OK instead of human people?
Ln125 – Looking at your code, there seems to be a concerning misunderstanding here.
summaryData$EQ <- summaryData$Brain*1.036*(10**-3)/(0.085*summaryData$Bodymass**0.775) #Following decasien, according to #Jerison, H. J. Evolution of the Brain and Intelligence (Academic, 1973).
What you are doing here is you are multiplying the brain volume as per Jerrison’s formula, but then you multiply by 10^3 – this is not needed, as the derived value is already a conversion from gr to mm3 (the 1.036 being the conversaion factor)
Ln 138 - Additionally, when obtaining ‘relative brain partition size’ the partition size is being divided by bodymass:
summaryData$ratioNeocortex <- summaryData$Neocortex/summaryData$Bodymass
This is also inappropriate, as in order to control for the allometry it is better normalized by the whole brain size. I.e. brain partitions scale allometrically to whole brain size, not body size (they scale to body size too, but as a consequence of scaling to brain size).
See for example Triki, Z., Granell-Ruiz, M., Fong, S., Amcoff, M., & Kolm, N. (2022, May 2). Brain morphology correlates of learning and cognitive flexibility in a fish species (Poecilia reticulata).
Another approach is to scale it to another (more conserved) brain area as in Vanier, D. R., Sherwood, C. C., & Smaers, J. B. (2019). Distinct Patterns of Hippocampal and Neocortical Evolution in Primates. Brain, behavior and evolution, 93(4), 171–181. https://doi.org/10.1159/000500625
Additional methods comment – Seeing that you are sacrificing huge portion of your sample size (301) in the model fitting (N=70, 69, 39), have you considered using multiple imputation technique to deal with missing values? See Todorov, 2021; Nakagawa, 2011
Todorov, O. S., Blomberg, S. P., Goswami, A., Sears, K., Drhlík, P., Peters, J., & Weisbecker, V. (2021). Testing hypotheses of marsupial brain size variation using phylogenetic multiple imputations and a Bayesian comparative framework. Proceedings of the Royal Society B, 288(1947), 20210394.
Nakagawa, Shinichi, and Robert P. Freckleton. "Model averaging, missing data and multiple imputation: a case study for behavioural ecology." Behavioral Ecology and Sociobiology 65.1 (2011): 103-116.
Ln190 – Re Ancestral diet reconstruction – this method has been shown to be very unreliable when there is absence of fossil data to ‘correct’ for estimations on the ancestral nodes. If you cannot obtain data on extinct primate’s diet (at least a few and I am sure there can be found in the literature) I would advise against the use of ancestral state reconstruction.
Ln206 – encephalization, not encephalic. Also re the use of EQ, you might want to check van Schaik, 2021, where they advise using caution when using EQ.
van Schaik C, P, Triki Z, Bshary R, Heldstab S, A: A Farewell to the Encephalization Quotient: A New Brain Size Measure for Comparative Primate Cognition. Brain Behav Evol 2021;96:1-12. doi: 10.1159/000517013
Ln234 – Multiple imputation can solve a lot of problems here
Ln319 – This can be rephrased.
Download the review https://doi.org/10.24072/pci.evolbiol.100548.rev15









