
How the analysis of the distribution of fitness effects can reveal novel insights onto the genetics of domestication
 based on reviews by Miguel de Navascués and 1 anonymous reviewer
based on reviews by Miguel de Navascués and 1 anonymous reviewer
Detection of Domestication Signals through the Analysis of the Full Distribution of Fitness Effects
Abstract
Recommendation: posted 14 March 2025, validated 15 March 2025
Fumagalli, M. (2025) How the analysis of the distribution of fitness effects can reveal novel insights onto the genetics of domestication. Peer Community in Evolutionary Biology, 100795. 10.24072/pci.evolbiol.100795
Recommendation
The joint full distribution of fitness effects (DEF) is an important indicator in population genetic studies, and its inference has been the subject of intense research [1]. However, we still lack a solid framework to estimate DFE under certain demographic conditions.
In this study, Castellano and colleagues propose to estimate the DFE by analysing the site frequency spectrum (SFS), and specifically develop a new approach for the joint DFE model inference [2]. The latter is based on the proportion of variants with divergent selection coefficients. Authors performed extensive simulations under models of domestication which is arguably one of the most crucial series of events in human evolution [3]. Domestication is associated with significant genetic costs in animals [4].
While DFE is typically estimated by contrasting SFS of silent and functional mutations [5], it has been recently suggested to use the joint SFS between domesticated and wild populations to estimate the DFE [6]. Authors build on this model and expand its parameterisation. Authors were able to dissect the impact of linked selection on inferred demographic history of wild and domesticated populations, with a robust estimation of the deleterious DFE.
There are still several limitations in the interpretation of DFE as, for instance, some selective sweeps can bias their estimates and some demographic scenarios are challenging to infer. Also, classic quantitative trait models should be evaluated as a complementary approach. Finally, the in silico predictions presented in this study could be validated by empirical scans on existing genomic data sets. Nevertheless, this study is an important contribution to our understanding on how demography, and domestication in particular, can affect variants under selection in recent evolutionary histories.
References
[1] Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8(8):610-618. https://doi.org/10.1038/nrg2146
[2] Castellano D, Vourlaki IT, Gutenkunst RN, Ramos-Onsins SE. Detection of Domestication Signals through the Analysis of the Full Distribution of Fitness Effects. BioRxiv 2025, ver.4 peer-reviewed and recommended by PCI Evol Biol https://doi.org/10.1101/2022.08.24.505198
[3] Frantz LAF, Bradley DG, Larson G, Orlando L. Animal domestication in the era of ancient genomics. Nat Rev Genet. 2020;21(8):449-460. https://doi.org/10.1038/s41576-020-0225-0
[4] Schubert M, Jónsson H, Chang D, et al. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc Natl Acad Sci U S A. 2014;111(52):E5661-E5669. https://doi.org/10.1073/pnas.1416991111
[5] Kousathanas A, Keightley PD. A comparison of models to infer the distribution of fitness effects of new mutations. Genetics. 2013;193(4):1197-1208. https://doi.org/10.1534/genetics.112.148023
[6] Huang X, Fortier AL, Coffman AJ, et al. Inferring Genome-Wide Correlations of Mutation Fitness Effects between Populations. Mol Biol Evol. 2021;38(10):4588-4602. https://doi.org/10.1093/molbev/msab162
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
SERO. was supported by grants AGL2016-78709-R (MEC, Spain) and PID2020-119255GB-I00 (MICINN, Spain), by the CERCA Programme/Generalitat de Catalunya and acknowledges financial support from the Spanish Ministry of Economy and Competitiveness, through the Severo Ochoa Programme for Centres of Excellence in R&D 2016-2019 and 2020-2023 (SEV-2015-0533, CEX2019-000917) and the European Regional Development Fund (ERDF). RNG and DC were supported by the National Institute of General Medical Sciences of the National Institutes of Health (R35GM149235 and R01GM127348 to RNG). ITV was supported by a predoctoral fellowship funded by MCIN/AEI/ 10.13039/501100011033 through the Grant BES-2017-081139 and by “ESF Investing in your future”. This research was also supported by NSF Grant No. PHY-1748958 and the Gordon and Betty Moore Foundation Grant No. 2919.02.
Reviewed by Miguel de Navascués , 27 Jan 2025
, 27 Jan 2025
The authors have thoroughly revised their manuscript in response to the suggestions from the previous review. The new version is significantly clearer, and the additional results complement the earlier work. I recommend the publication of this manuscript.
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2022.08.24.505198
Version of the preprint: 3
Author's Reply, 09 Jan 2025
Decision by Matteo Fumagalli , posted 25 Jun 2024, validated 25 Jun 2024
, posted 25 Jun 2024, validated 25 Jun 2024
Dear authors,
thank you for your submission. Your study has been evaluated by two experts in the field. Both of them find it of relevance and importance, as your results provide a good contribution to the field. The focus on domestication has also been appreciated.
However, reviewers point to several concerns with your submission. Both indicate a lack of details in the description of the methods, especially with the new approach to infer demography and selection. The code provided on github is not sufficiently documented. There are also some concerns over the interpretation of some results, where the reviewers appear to be less optimistic than you on the performance of your inferences. I also find Figure 3 to be too cluttered.
While the study has merit, I encourage the authors to carefully consider all the points raised by the reviewers to clarify your text accordingly.
Matteo
Reviewed by anonymous reviewer 1, 15 Jun 2024
Title and abstract
Does the title clearly reflect the content of the article? Yes
Does the abstract present the main findings of the study? Yes
Introduction
Are the research questions/hypotheses/predictions clearly presented? Yes
Does the introduction build on relevant research in the field?Yes, but it lacks some citations. I have added those to my review.
Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? No (please explain) The methods are not presented in detail. I have specified that in my review.
Are the methods and statistical analyses appropriate and well described? No (please explain)
Results
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? I don’t know
Are the results described and interpreted correctly? Yes
Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? Yes
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? Yes
Reviewed by Miguel de Navascués , 14 Jun 2024
, 14 Jun 2024
Title and abstract
- Does the title clearly reflect the content of the article? No: Title can be more clear (discussed below)
- Does the abstract present the main findings of the study? Yes
Introduction
- Are the research questions/hypotheses/predictions clearly presented? No: Objectives of the study could be presented in a more clear way (discussed below)
- Does the introduction build on relevant research in the field? Yes
Materials and methods
- Are the methods and analyses sufficiently detailed to allow replication by other researchers? No: Code is provided to replicate simulations and analyses, but description is insufficient to be understood (discussed below)
- Are the methods and statistical analyses appropriate and well described? No: Methods are appropriate as far as I can tell, but description is insufficient (discussed below)
Results
- In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? I don’t know
- Are the results described and interpreted correctly? Yes
Discussion
- Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? Yes
- Are the conclusions adequately supported by the results (without overstating the implications of the findings)? Yes
In this work, the authors address the performance of statistical methods to infer demography and selection (distribution of fitness effects, DFE) under an evolutionary scenario representing a domestication process. Some of the methods evaluated are widely used in the community, and one is a newly developed approach that accounts for the change of selection regime in the domesticated population. While the results reflect previously published evaluations of the negative effects of linked selection on demographic inference and the challenges of disentangling selection and demographic effects, the work merits attention for its specific focus on domestication and the development of a new approach. Contrary to the opinion of previous reviewers, I argue that the domestication scenario is a societally important evolutionary process with broad interest. Moreover, a scenario of divergent populations with one undergoing a bottleneck and a change of selection regime may also be relevant in other contexts, such as the case of invasive species. Unfortunately, the text presenting the work remains unclear, particularly regarding the description of the new approach. Below, I provide a list of points (roughly ordered from "concerns" to "suggestions") that need to be addressed before the work can be recommended.
- The description of the new approach (lines 275-334) lacks sufficient detail for the reader to understand the new development and its implementation. First, little explanation is provided about the previous developments upon which the new approach is based. There is just a sentence citing four works, leaving the reader to decipher why these works are cited, what key elements they provide for the new method, and how they were used. I assume the work builds upon the developments of Huang et al. 2021, and that Jerison et al. 2014 and Ragsdale et al. 2016 are cited for the concept of joint DFE. Assuming this is correct, the text should briefly describe the approach in Huang et al. 2021. Jerison et al. 2014 might fit better in the introduction, and the relevance of Ragsdale et al. 2016 (which focuses on the joint DFE of triallelic sites) is unclear. If my assumption is incorrect, the text should be revised to clarify this for the reader.
The text describes the proportions of different types of sites under selection used in the new approach. However, it is unclear how these proportions affect the site frequency spectrum (SFS) and how they are applied in practice for the inference. The authors refer readers to the code implementing the model without providing a mathematical description of it. It is imperative that the article clearly states how these proportions of different sites under selection modify the equations/algorithms (presumably from Huang et al. 2021) that provide the expected SFS. Otherwise, understanding the model would require reverse engineering from the code, which is unreasonable. Describing the work upon which the new model is built will provide the necessary elements for explaining the new model. - The description of the mutation process simulation is unclear. A more detailed description is needed on how the location of different types of sites is “randomly precalculated” and the proportion of different types of sites (m1, m2, etc.) in the genome (authors only report pc for the simulations). It might also be useful for the authors to explicitly explain whether the different proportions of sites are predetermined as fixed proportions or if each site type is randomly assigned and how many realized mutations of each type occur in the simulation. Clarify whether pc( $p_c$ ) is a probability or a proportion and how that translates to the actual proportion of mutations involved in domestication.
- The notation throughout the paper requires thorough revision. While some remarks on notation might be considered arbitrary conventions, others are necessary to avoid confusion. Symbols that represent quantities or variables should be in italics while labels (“e” for effective, “d” for deleterious, etc.) should be in roman (“upright”): several indexes in the notation used should be in roman: pc, Ne, sd, etc. I suggest to avoid the use of letter “x” as symbol for multiplication: no symbol (multiplication between two symbols) or the multiplication symbol (×) is a less ambiguous. I suggest to avoid using operators (“+”, ”-”) as labels when they can create confusion in equations (e.g. line 286), or, if kept, use a less confusing arrangement, for instance pw+ instead of p+w (as in pc+). I think that three letters (e.g. p, q and g or some other choice) might be even a simpler (thus better) option to pc, pc+ and pw+. In line 266, I suggest to replace n for i or to clarify the relationship between n and i. I suggest to use “%” only for percentages, i.e. express selection coefficients as -0.01 instead of -1%, express probabilities as numbers between 0 and 1 (note, it is not the same that each locus has a probability 0.25 of changing their selection regime that 25% of loci change their selection regime). It would be good to be more consistent with the notation throughout the manuscript: in the methods Sb and Sd are defined respect to Ne (line 222, Sd=2Nesd); however, in the simulation model and the inference model described in figure 1, Ne is not defined (does it correspond to Na?, to New?); then, γ is used for the population (Na) scaled selection coefficients instead of S which can lead to further confusion. The authors try to clarify some of this in lines 353-355 but it is still unclear the relationship between parameters because there is a lack of explicit definition of equivalences (4Nes=2Nas?). I would suggest to use a uniform notation throughout the paper and provide a complete description of the relationships between parameters. If there are not equivalences between models (e.g. simulation model and inference model) I think that author should state clearly so when comparing those parameters. Their lack of equivalence does not come from scaling respect to Ne or Na, because Na is defined in both models: I think authors should rescale, if necessary, to the same reference N to be consistent. Regarding the equivalence to SLiM notation, the explanation is only relevant for readers that will use the simulation code and (in my opinion) that explanation should be a comment within the code, not the main text. This will simplify the text for the general reader that does not need those technical details for understanding the analysis.
- In several parts of the text the description of reality, models and inferences is sometimes confusing, attributing model features to reality or describing inference results as processes. For instance, the abstract states that “Artificial selection can alter the selection coefficients”. A change in the selection regime can be modelled as changes of selection coefficients, as done by the authors, but it can also be modelled as a new optimum for a quantitative trait determined by a set of loci whose effects on fitness is not determined by a selection coefficient but by their effect on the trait and the genetic background they are in. There is not such a things as selection coefficients for real alleles in a domesticated population, only models that try to describe the actual process in a useful way. In lines 468-469 authors state that strong selection “erase[s] the demographic history” and “recreate[s] large recent population expansions”. The presence of strong selection does not change demography (in the simulated model of this work), but it can challenge its inference: I suggest the authors to use words that convey that idea (e.g. “erases the signal of demography” and “generates genetic diversity patterns similar to recent population expansions”). In line 449, is it the “ demographic history” or “the inferred demographic history”? I suggest to revise the whole text to this regards.
- Results and discussion section introduce analysis and methodological details that should have been described in the methods section. The description of genetic diversity presented in lines 652-683 is not described in the methods and it is not described how the different is calculated. In line 381 a key information about the codominance of simulated mutations if given instead of being stated in the methods. Also, methods do not clearly state from which data all the calculation are produced (samples of individuals of the last generation?)
- Regarding the discussion of the results from the estimation of the marginal DFE the authors speculate about domesticated populations having higher number of advantageous mutations due to migration. I do not understand this reasoning because migration should also introduce deleterious mutations. I think the authors should provide a more clear explanation of their reasoning and they should backup their statement by looking at the simulated data the actual proportions of advantageous mutations of their models.
- I suggest to remove altogether the paragraph about polygenic adaptation (lines 83-96). I do not see the relevance. The discussion of considering the fitness as a trait is also misleading, the type of dynamics that selection of quantitative traits can have could be very different to what it is modelled in this work. An allele of a quantitative trait locus is not deleterious or beneficial per se, it would be positively selected or negatively selected depending on the diversity of the individual genomes in which it occurs and on the whole genomic composition of the population,and this can change through time even if the phenotypic optimum remains constant.
In the same line, I suggest to replace “considers polygenic adaptation (considering fitness as a trait)” for “considers multilocus adaptation” - I suggest to revise the title, which is misleading. Current version seems to state that “detection of domestication signals” is done “using simulations”. I would suggest removing “using simulations” because keeping that in the title would lead to a more cumbersome explanation: e.g. “Evaluation of methods to detect … using simulations”.
In the same line, I suggest to revise the first sentence of the materials and methods (line 170). A “simulation analysis of the domestication process” does not seems a proper description of what it was done. The simulation are used to evaluate the performance of several inference methods, there is no analysis of the domestication process.
And again, in the conclusions (line 703), the simulations “provide valuables insights” on method performance, not on demography and adaptation in the context of domestication. By using a simulation model, the genetic dynamics of domestication are assumed to follow specific processes determined by the authors vision of domestication. As discussed above, there is some confusion between reality and model/inferences in the discourse of the authors. - The authors share the code used in their work through a GitHub repository. This is good but it must be noted that this does not completely comply with FAIR criteria. Research data repositories should assure the permanent availability of the shared data. A GitHub project could be removed by decision of the project owner or GitHub could cease to exists (GitHub has not vocation of being a permanent repository; therefore, there are not provisions to assure the persistence of research data contained in their projects). I strongly encourage the authors to deposit their code in a proper research data repository. I suggest, as fast and easy solution, to use Zenodo, as it has tools to link with GitHub projects.
- Line 586. It is unclear what is meant with “easier”. I think the authors mean that the inference of deleterious DFE is robust to demographic model misspecification, but in any case the sentence would benefit from some revision.
- Line 124. Does not the work by Huang et al. 2021 provide a method to detect differences in DFE between closely related populations?
- In the abstract, the objectives of the study, as presented in the first four sentences, are vague. First, there is a mention to “genetic consequences”, which could mean anything. Then there is a confusing sentence mixing the domestication process with one model to describe it (see previous comment) and the next sentence starts with “To investigate this” but it seems unclear to me what “this” refers to. I suggest to revise this sentences.
- lines 118-120. Please develop or give some references, it seems unclear to me to what kind of study this sentence is referring.
- lines 25-26: I suggest to remove (or modify) “such as the shape and strength of the deleterious DFE”. First, a distribution has no “strength”, I think authors mean the “mean” of the distribution. Then, the type of assumed distribution is not mentioned, so mentioning the “shape” parameter does not make much sense here. Also, note that this is also a confusion between reality and the model (as discussed previously), DFE have a gamma distribution in (some) models.
- line 23: replace “fluctuating” with “changing” (or equivalent). There is no fluctuating selection in the models considered in this work.
- line246: What is it meant with “disposition”? “arrangement”?
- line 143: I suggest to replace “SLiM simulations (ref)” for “forward-in-time simulations” (here the specific software used in not relevant and the reader cannot understand the type of simulation if they don't know about SLiM)
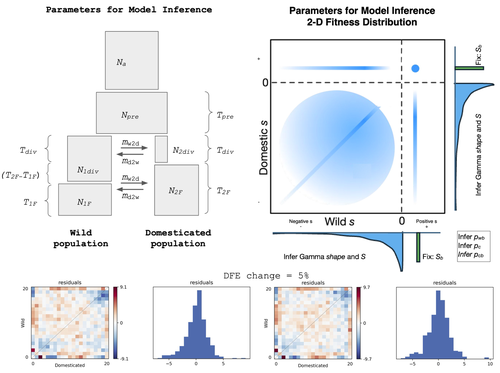
- figue 1: I suggest to remove “=1” from “Na=1” or explain what does it mean.
- lines 111-113. This explanation might be better placed in the first mention of the terms 1D-SFS and 2D-SFS.
- lines 693-694: Maybe this can be clarified or developed more. I am unsure if the authors are referring to already existing methods (which ones) or methods that should be adapted or developed.
- line 51: I might be missing some subtle meaning here but, isn’t “artificial selection” always “human-induced”?
- line 611: typo, it should be “out” not “our”
- line 223-224: format of references should be revised
- line 271: “R” should be removed
- line 847: incomplete reference.










