
Is genetic diversity enhanced by a supergene?
Balancing selection at a wing pattern locus is associated with major shifts in genome-wide patterns of diversity and gene flow in a butterfly
Abstract
Recommendation: posted 29 March 2023, validated 30 March 2023
Jiggins, C. (2023) Is genetic diversity enhanced by a supergene?. Peer Community in Evolutionary Biology, 100522. https://doi.org/10.24072/pci.evolbiol.100522
Recommendation
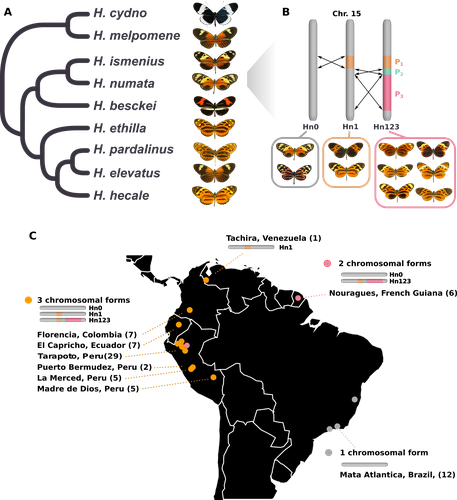
The butterfly species Heliconius numata has a remarkable wing pattern polymorphism, with multiple pattern morphs all controlled by a single genetic locus, which harbours multiple inversions. Each morph is a near-perfect mimic of a species in the fairly distantly related genus of butterflies, Melinaea.
The article by Rodríguez de Cara et al (2023) argues that the balanced polymorphism at this single wing patterning locus actually has a major effect on genetic diversity across the whole genome. First, polymorphic populations within H. numata are more dioverse than those without polymorphism. Second, H. numata is more genetically diverse than other related species and finally reconstruction of historical demography suggests that there has been a recent increase in effective population size, putatively associated with the acquisition of the supergene polymorphism. The supergene itself generates disassortative mating, such that morphs prefer to mate with others dissimilar to themselves - in this way it is similar to mechanisms for preventing inbreeding such as self-incompatibility loci in plants. This provides a potential mechanism whereby non-random mating patterns could increase effective population size. The authors also explore this mechanism using forward simulations, and show that mating patterns at a single locus can influence linked genetic diversity over a large scale.
Overall, this is an intriguing study, which suggests a far more widespread genetic impact of a single locus than might be expected. There are interesting parallels with mechanisms of inbreeding prevention in plants, such as the Pin/Thrum polymorphism in Primula, which also rely on mating patterns determined by a single locus but presumably also influence genetic diversity genome-wide by promoting outbreeding.
REFERENCES
Rodríguez de Cara MÁ, Jay P, Rougemont Q, Chouteau M, Whibley A, Huber B, Piron-Prunier F, Ramos RR, Freitas AVL, Salazar C, Silva-Brandão KL, Torres TT, Joron M (2023) Balancing selection at a wing pattern locus is associated with major shifts in genome-wide patterns of diversity and gene flow. bioRxiv, 2021.09.29.462348, ver. 3 peer-reviewed and recommended by Peer Community in Evolutionary Biology. https://doi.org/10.1101/2021.09.29.462348
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
This work was funded by grants Hybevol (ANR-12-JSV7-0005) and Supergene (ANR-18-CE020019-01) from the Agence Nationale de la Recherche and European Research Council Grant MimEvol (StG-243179).
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/2021.09.29.462348
Version of the preprint: 2
Author's Reply, 27 Feb 2023
Decision by Chris Jiggins, posted 03 Jan 2023, validated 04 Jan 2023
This revision is a great improvement and I agree with the reviewer that it addresses many of the previous concerns. The Dadi reconstruction showing increase in Ne in polymorphic numata is a nice addition.
The simulations are also a welcome addition, but need better description and presentation. In Figure 4 the legend is incomplete - I assume the colours represent the migration parameter but this is not stated. Why not put assortative - disassortative mating on a single panel as this is a continuum. It would also be helpful to outline the justification for the modelling of local adaptation with gene flow - is that based on observed spatial heterogeneity in mimetic models in H. numata?
I also agree with the reviewer that random mating should be the null hypothesis - there isnt any Heliconius where assortative mating has been demonstrated between sympatric morphs as far as I am aware, so not clear that the null hypothesis should be assortative mating. From the figure, it would seem that dissassortative mating does not make much difference to diversity as compared to random mating, it is rather assortative mating that reduces genomic variation? The authors need to clarify the interpretation here. Nonetheless, the simulations do show that mating patterns at a single locus can significantly affect linked diversity, so provide some support for the overall hypothesis. I also found myself wondering whether unlinked diversity would be similarly influenced - as only a 1MB chromosome was simulated. The authors could perhaps address this in the discussion.
I therefore request the authors to address these concerns and those listed in the review. I do not anticipate that any further review is necessary.
Reviewed by Christelle Fraïsse , 21 Dec 2022
, 21 Dec 2022
I am delighted with this new version and the comprehensive reply letter. The author made substantial efforts to address my and other reviewers’ concerns, and they performed additional analyses:
1) perform two demographic analyses to estimate the timing of Ne changes explicitly.
2) perform extensive individual-based forward simulations to confront their observations with predictions under different models.
I also liked very much that the authors were more cautious in their interpretation. They made the necessary changes in the abstract, results and discussion.
I still have a few suggestions below to handle before publication; they are minor. Therefore, I fully support the release of their great study in PCI.
Note that line numbers refer to the “tracked changes” version.
Forward simulations:
● Material and methods:
- L288 and following: Justify in biological grounds the setting of your simulations (stepping stone model, number of morphs = 5, 2 or 3 morphs have a fitness advantage per population). Is this setting in agreement with what is known from the system? How are the morphs initially distributed across the demes?
- L309 – 327: your parametrization of the strength of assortative/disassortative mating is confusing. I would rephrase it in terms of the probability of mating with a different morph, where a value of 0 = assortative mating, of 1 = disassortative mating and 0.5 = random mating, as you did in Figure 4. It will be more intuitive to interpret.
- Moreover, I need clarification on why you tested only values from 0 to 0.5 (in your parametrization) in both the dis-assortative and assortative mating. A value of 1 is omitted, while this corresponds to a “random mating model” (as far as I understand). So in the text, you say that you do not test for “random mating” (which is the null model against which the disassortative model should be tested against). But then, in Figure 4 it is indicated that you tested a case “equivalent to random mating”. Sorry for the confusion. Could you clarify please?
● Results (from L396 to 404):
- I am not sure to understand why you highlighted the difference between the case of assortative mating vs disassortative mating (average piS of 0.011 vs 0.0145). Is assortative mating the common reproduction mode when the supergene is absent in these butterflies? In addition to this comparison, could you please briefly describe the difference with the simulations under random mating?
- You could put the weak difference obtained in the simulations in perspective with the observed differences (<0.01 for the Brasilian H. numata vs >0.025 for the Amazonian H. numata). This hints at the minor contribution of assortative mating to the neutral genetic diversity compared to other factors, such as the migration rate between demes or founder events. This can be highlighted, for example, along lines 490 in the Discussion.
- Figure S5: It is unclear which simulations were pooled under the label “assortative mating” and under “disassortative mating”. All parameter combinations? It would be clearer to color the points according to the strength of the assortative (or disassortative) mating.
Minor suggestions to handle before publication:
● L188: “(I)” → “(i)”.
● L227: typo in “at which LD decay[s]”.
● L235: please, add “i.e.,” before “migration rate times the duration of the migration band”.
● L276: please, add after “controlled by parameters s1 and s2”, the new time parameters (Tp1 and Tp2), which indicate the time of exponential change.
● L285: “and only models with the lowest” → do you mean the “run” with lowest… ?
● L302: typo in “with a given [set of] deleterious recessive mutation[s], generating overdominance at [these] loci”.
● L306:
- remove “reduction” in “fitness reduction varied”.
- “fitness of zero for migrants in a deme” → “null fitness for non-locally adapted individuals”.
- remove the “s” in deme[ ].
● L311:
- add after “complete disassortative mating”, a short explanation of what it means, such as “where a given individual mates only with different morphs”.
- “no mating weight” is unclear; please, consider replacing with “random mating”.
● L325: please, remove “no disassortative mating but”.
● L326: please, remove “(or no assortative mating)”.
● L349: typo in “other Amazonian population[s]”.
● L390: you could also mention the inconsistency of the point estimate of Ne for the peruvian H. numata population in the two inferences (with H. pardalinus or with brazilian H. numata). However, they both show a large Ne for the peruvian H. numata.
● L442: I would replace “coincide” with “coincide in time”.
● L479: I would replace “translate into an effect on effective migration genomewide” with “translate into a reduction of the effective migration genomewide”.
● L489: typo in “results also suggest[ ]”.
● L722: typo in “a[d]mixture”.
● L730: a bracket is missing between “numata” and “populations”.
● Figure 4:
- title: “and mating region” should be removed or rephrased, because it is confusing as it states.
- I am not sure to understand what the left number in brackets means. Is 0.5 random mating? Because you state it is neither “assortative mating”, nor “assortative mating”.
- In the same line, I do not understand the difference between the set of parameters n°7-9 in panel A vs n°1-3 in panel B.
- Why the right number in brackets is increasing in panel A, while it is decreasing in panel B?
- Could you please indicate the meaning of the different colors?
- L761: “10 populations” → do you mean 10 replicates?
- L763: please, add “strength of” before “local adaptation”.
- L767: typo in “in the brack[et] mean[s]”.
- L768: typo in “in a deme[ ]”.
Below, line numbers from the biorxiv version.
● Table 1:
- L715: typo in “Biological parameter assum[]ed”.
- L716: I would remove “, and descending populations 1 and 2”.
- L725: “for” → “four”.
● Figure S1: typo in “H. numata individual[s]”, “microsatel[l]ites”, “20 STRUCTURE run[s]”.
● Figure S2: label the x axis of the left bottom residual plot.
● Table S7: a note beside the table could indicate that the optimization of the logL was sometimes unsuccessful as some nested models have a higher likelihood than their general version (for example, SI2NG is much less likely than SI2N, while it should be at least as likely because it is the same model but with an extra parameter).
https://doi.org/10.24072/pci.evolbiol.100522.rev21Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2021.09.29.462348
Version of the preprint: 1
Author's Reply, 15 Nov 2022
Decision by Chris Jiggins, posted 30 Dec 2021
This is an exciting paper that links mimicry polymorphism to broad genome-wide population genetic parameters. However, all three reviewers raise concerns about the manuscript as it stands. The general issue is that with a sample size of 1 there is only rather tentative evidence to link the increase in Ne with supergene formation. This is clearly acknowledged in the Discussion section but not really reflected in the title and abstract and general framing of the paper. One reviewer suggests additional analyses to try and infer the timing of Ne changes relative to supergene introgression, which would provide further evidence for a causal link. An alternative approach might be to use simulations to estimate the increase in Ne expected for a given level of disassortative mating and balancing selection (these parameters are presumably reasonably well known in H. numata) - comparison of empirical and theoretical expectations might help support the hypothesis.
It is worth noting that H. melpomene shows a similar difference between Amazonian and Atlantic forest populations, with Atlantic populations far less diverse (4 H. m. nanna samples were published here:
Belleghem, S. M. V. et al. Patterns of Z chromosome divergence among Heliconius species highlight the importance of historical demography. Molecular Ecology 27, 3852–3872 (2018).). This somewhat undermines the argument that this difference is due to the supergene in H. numata, but analysis of the melpomene population data would provide a contrast that may support the proposed hypothesis if a much greater Amazon/Atlantic difference is seen in H. numata.
Overall there are also a large number of smaller comments that need addressing.
Regarding the broad conclusions the supergene hypothesis either needs to be reduced in prominence through the text, or some additional analyses conducted to further support the hypothesis (the latter would be much preferable).
Reviewed by anonymous reviewer 1, 15 Dec 2021
Reviewed by Christelle Fraïsse , 23 Dec 2021
, 23 Dec 2021
In this manuscript, de Cara and collaborators investigate the genome-wide effect of a supergene controlling wing patterns in Heliconius butterflies. Based on whole-genome resequencing, they showed that the Amazonian populations of H. numata are more diverse and less structured than all other taxa they investigated. These populations are also the only ones polymorphic for the supergene. The authors, therefore, hypothesize that disassortative mating following the onset of polymorphism through adaptive introgression is responsible for the enhanced diversity observed in H. numata Amazonian populations.
The main results that adaptive introgression can affect population demography (i.e. gene flow and effective sizes) due to change in the mating system is appealing. And I agree with the authors that few studies carefully investigated this effect, making the present work valuable. That being said, I am not entirely convinced that the authors have clearly demonstrated the connection between polymorphism at the supergene and enhanced diversity. Mainly, I think that the demographic analyses should be strengthened. Please, see the detailed comments hereafter.
1. Demographic inferences – alternatives to G-PhoCS:
The main results of this work are based on the comparison of closely-related populations differing at a trait affecting genome-wide diversity, coupled with knowledge of when the differences evolved (L70-71). More precisely, the supergene formation should precede the change in demography. Therefore, it is essential to carefully estimate the timing of population size (Ne) and gene flow (M) changes. From this point of view, G-PhoCS does not seem to be the most appropriate method (see below), so I think a different type of demographic inferences should complement it.
First, G-PhoCS assumes that Ne is constant along branches of the phylogeny, and so Ne can change only when the ancestral population diverge. Moreover, the method cannot capture bottlenecks or size expansions on individual branches. The changes in population sizes (and their timing) should be tested explicitly in the present study.
Second, in the original G-PhoCS paper (Gronau et al. 2011), the authors tested the ability of their method to estimate Ne and M accurately based on simulations. They found that the method has limited power given the features of their data. This should encourage the authors to perform similar validation analyses in the present study.
The third limit of G-PhoCS is its computational burden which led the authors only to use two diploid genomes per taxa.
I understand that the advantage of G-PhoCS is to reconstruct the history of many species along a phylogeny, where other demographic inference methods are mainly applied to 2-population comparisons. Considering closely-related lineages in the inference is particularly important if genetic exchanges are pervasive among the taxa. Yet this is not the case of H. numata, which is only connected to H. pardalinus (or its ancestor) based on results in Table S6.
=> For all these reasons, I think it would be helpful to strengthen the demographic results by running a different method that can explicitly model temporal changes in Ne and M and use data from more than two samples. Such methods are implemented in programs like dadi (Gutenkunst et al. 2009: 10.1371/journal.pgen.1000695), moments (Jouganous et al. 2017: 10.1534/genetics.117.200493), FastSimCoal (Excoffier et al. 2011: 10.1093/bioinformatics/btr124) or DILS (Fraïsse et al. 2021: 10.1111/1755-0998.13323). The 2-population versions of these programs could be used (Amazonian vs Atlantic H. numata), or a 3-population version if H. pardalinus is to be considered.
Alternatively, the authors could reconstruct the changes in Ne through time for each population using PSMC-like methods (e.g. Li & Durbin 2011: 10.1038/nature10231), but then, migration has to be neglected.
2. Demographic inferences - other comments
- As far as I understand, what is expected in terms of Ne is an increase in the polymorphic populations (Amazonian H. numata), which agrees with the inferences (Figure 3). However, connecting the “polymorphic / monomorphic” status with genetic diversity and effective size is not so obvious. Indeed, G-PhoCS infers a demographic expansion in H. elevatus, while this species is monomorphic. Moreover, the diversity difference between the two H. numata populations is partly explained by the decrease in the size of the Atlantic populations (Figure 3).
- From the PCA (Figure 2), three genetic clusters make up H. numata (Atlantic, Amazonia, French Guiana) with similar variance explained along each axis. I wondered whether this could inflate the effective size of the Amazonian H. numata populations inferred with G-PhoCS. Given the population structure (even if weak), it may be worth applying the demographic analyses without the French Guiana samples.
- G-PhoCS assumes no intralocus recombination but free interlocus recombination. Do the filters given on L205 (“4092 genomic regions, each 1kb in length and spaced at approximately 30kb intervals”) comply with these assumptions?
- On L200, it is stated that the “inferences are conditioned on a given population phylogeny”. I wondered how robust is the phylogeny used in G-PhoCS (Figure 3)?
- Could you please justify what the criteria of Freedman (L213) is based on? It is not obvious why the migration rate threshold of “0.03 with posterior probability larger than 0.5” is meaningful here.
3. Supergene polymorphism:
- I think it would be helpful to be more precise regarding what is expected for the different types of configurations. Are differences in genetic diversity expected: i) between polymorphic populations with two vs three configurations?; ii) between monomorphic populations with Hn0 (Brazil) vs Hn1 (Venezuela) configurations?
- An interesting point not discussed by the authors is the presence of a monomorphic H. numata population (carrying the inversion Hn1) in Tachira (Venezuela). This sample (n=1) is not presented in Figure 3, so we cannot compare its diversity (pi) with that of the other H. numata populations. If the authors decide to go with PSMC as an alternative demographic method, they can even include the Tachira sample in the analysis to estimate its temporal Ne changes.
4. Confounding factors:
- Given the phylogenetic proximity of the taxa considered, I suspect that they should share similar geographic barriers or have a similar dispersal rate. Still, this may not be true. And if differences exist between taxa, caution is required to interpret the isolation by distance patterns shown in Figure 2C. Typically, if H. numata has higher dispersal capacities than the other taxa and if a geographic barrier exists in the disjunct species range (South East of Brazil, Figure S2), then Figure 2C could be explained without the need of invoking the supergene effects. Could the authors comment on that?
- In Figure 2C, it seems that two Fst values are not depicted based on Table S4: between H. pardalinus butleri vs sergestus (Fst=0.30574, 30 km) and between H. pardalinus sergestus vs Ssp (Fst=0.30155, 430 km). These two values are outliers (i.e. strong differentiation at small distances). Could the authors explain why they were removed from the figure? Do they correspond to subspecies? At least, an explanation has to be indicated in the legend of Figure 2C.
5. Minor comments
- L41: “they show the highest [...] demographic estimates”. Please, reformulate and specify what are the demographic estimates.
- L190: “Scaffolds carrying the supergene rearrangements (Hmel215006 to Hmel215028) were excluded”. Does this correspond to the whole chromosome 15 or only to the scaffolds of chromosome 15 that carry the supergene?
- L275: “which contrasts with the low diversity found in the most closely related taxa such as H. ismenius or H. besckei”. I think that H. besckei does not appear anywhere in the results.
- L282: “the distribution of parameters across lineages”. The wording is a bit unclear; please reformulate.
- L349-351: this sentence sounds redundant with L340-343.
- Figure 1C: it was unclear whether the colour code (orange, pink, grey) refers to the number of chromosomal configurations. If yes, the Tachira sample should be depicted in grey.
- Figure 2B: maybe, indicate “Brazil (Atlantic)” instead of “Atlantic” in the legend.
- Figure 3A: a space is missing between “H.” and the rest of the name in the legend. Moreover, there is a typo in “Numata French Guiana PR” (I think “PR” should be “FG”).
- Figure 3B: there is an extra “s” in “Population names indicate[s]”, and there is a typo in “[showing] that Amazonian”.
- Figure S1: an “s” is missing in “20 STRUCTURE run[]”, and “reps” should be replaced by “replicates”.
- Table S4: dxy was calculated, but it was not used in the manuscript. Could the authors comment on that?
- Table S5: it seems that some values do not fit with the ones reported in Figure 3B.
- Table S6: it seems that the “probability that the estimated total migration was greater than 0.03” column is absent.
- Text S1: the “Analyses of the slope of Fst versus distance as measured in km” results are hard to follow. It would be clearer if the expectations for each test were stated before the results.
- Text S1: the “Testing for an effect of the inversion on population differentiation” results are hard to follow. Maybe add a sentence that clearly says if results with vs without chromosome 15 were the same or not.
https://doi.org/10.24072/pci.evolbiol.100522.rev12Reviewed by anonymous reviewer 2, 22 Dec 2021
This is a short article describing the genetic diversity of populations of Helionius numata, with comparisons with that of other Heliconius butterfly species.
The main finding seems that the H. numata species is divided into two populations:
A large population in Amazonia, which includes the samples taken from the Andean foothills (the vast majority of samples in the study) and samples from a locality in French Guiana; this population has very high genetic diversity and low isolation by distance.
A population in the Mata Atlântica region, represented by four samples, which has much lower genetic diversity.
The authors compare the genetic diversity of these two populations with that of populations of other Heliconius species, and include a model of the evolution of these species using the programme G-PhoCS. These analyses suggest that the Amazonian population does indeed have higher genetic diversity than the ancestral state.
I have two major reservations regarding this article.
First, the sampling strategy is extremely unbalanced: the genetic diversity of the Amazonian population is estimated with many samples from numerous places, whereas the diversity of the samples are estimated from only four samples from a single locality. The authors give excellent evidence that the genetic diversity of Amazonian populations is high. However, without a larger sampling breadth for the Mata Atlântica region, it is impossible to know whether the sampled population is representative of the entire region. As far as we know, the Atlantic forests of Brazil are comparatively more highly fragmented than the Amazonian rainforest - a recent bottleneck of the sampled population cannot be ruled out.
The second major reservation is the interpretation of the genetic diversity as being caused by disassortative mating as a result of dominance-selection regime of the mimicry polymorphism. The authors do not show that the genetic diversity observed is higher than what would be expected under a very large population size and random mating. Because of that, the main claim (as plausible as it is) seems too forceful in the way it is expressed in the Title, Abstract, Introduction and Discussion. Saying that, we do agree that the authors should discuss the possible importance of disassortative mating in the Discussion section.
Minor comments:
The Methods and Results are written well, but lack preciseness in places.
in line 222 and line 226, the authors should mention the number of individuals in each population (n=XX)
For the FST analysis (in line 234, Figure 2C and the relevant part of the Methods section), it is unclear what the authors are doing: which populations are compared with which populations?
In Figure 3A, it is not very clear which population is the numata one from Mata Atlântica. This is a general trend across the figures - the labels and biological categories are difficult to track across figures.
https://doi.org/10.24072/pci.evolbiol.100522.rev13









