Despite reproductive interference, the net outcome of reproductive interactions among spider mite species is not necessarily costly
Salomé H. Clemente, Inês Santos, Rita Ponce, Leonor R. Rodrigues, Susana A. M. Varela and Sara Magalhães
10.1101/113274
The pros and cons of mating with strangers
Recommended by Vincent Calcagno based on reviews by Joël Meunier and Michael D Greenfield
Interspecific matings are by definition rare events in nature, but when they occur they can be very important, and not only because they might condition gene flow between species. Even when such matings have no genetic consequence, for instance if they do not yield any fertile hybrid offspring, they can still have an impact on the population dynamics of the species involved [1]. Such atypical pairings between heterospecific partners are usually regarded as detrimental or undesired; as they interfere with the occurrence or success of intraspecific matings, they are expected to cause a decline in absolute fitness.
The story is not always so simple however, and it might all depend on the timing of events and on the identity of the partners. Using the herbivorous mite Tetranychus urticae as a model, Clemente et al. [2] experimentally arranged matings with two other Tetranychus species that commonly share the same host plants as T. urticae. They carefully controlled the history of events: heterospecific matings could occur just before, just after, 24h before, or 24h after, a conspecific mating. Interestingly, the oviposition rate (total fecundity) of females was increased when mating with a heterospecific individual. This suggests that heterospecic sperm can stimulate oogenesis just as conspecific sperm does. Such a positive effect was observed for matings involving T. ludeni females and T. urticae males, but a negative effect is found in the interaction with T. evansi. Sex-ratio (fertilization success in those species) could also be impacted but, unlike fertilization, this occurred when the mating events were distant in time. This is is at odds with what is observed in conspecific matings, where sperm displacement occurs only if mating events are temporally close. Overall, the effects of heterospecific mating were quite variable and it is challenging to predict a single, general, effect of interspecific matings. The net effect will likely be context-dependent, depending on the relative frequency of the difference mating sequences and on how fecundity and sex-ratio contribute to overall fitness, both aspect strongly influenced by the population dynamics and structure.
References
[1] Gröning J. & Hochkirch A. 2008. Reproductive interference between animal species. The Quarterly Review of Biology 83: 257-282. doi: 10.1086/590510
[2] Clemente SH, Santos I, Ponce AR, Rodrigues LR, Varela SAM & Magalhaes S. 2017 Despite reproductive interference, the net outcome of reproductive interactions among spider mite species is not necessarily costly. bioRxiv 113274, ver. 4 of the 30th of June 2017. doi: 10.1101/113274
| Despite reproductive interference, the net outcome of reproductive interactions among spider mite species is not necessarily costly | Salomé H. Clemente, Inês Santos, Rita Ponce, Leonor R. Rodrigues, Susana A. M. Varela and Sara Magalhães | Reproductive interference is considered a strong ecological force, potentially leading to species exclusion. This supposes that the net effect of reproductive interactions is strongly negative for one of the species involved. Testing this requires... |  | Behavior & Social Evolution, Evolutionary Ecology, Species interactions | Vincent Calcagno | | 2017-03-06 11:48:08 | View |
Evolution of resistance to single and combined floral phytochemicals by a bumble bee parasite
Palmer-Young EC, Sadd BM, Adler LS
10.1111/jeb.13002
The medicinal value of phytochemicals is hindered by pathogen evolution of resistance
Recommended by Alison Duncan and Sara Magalhaes
As plants cannot run from their enemies, natural selection has favoured the evolution of diverse chemical compounds (phytochemicals) to protect them against herbivores and pathogens. This provides an opportunity for plant feeders to exploit these compounds to combat their own enemies. Indeed, it is widely known that herbivores use such compounds as protection against predators [1]. Recently, this reasoning has been extended to pathogens, and elegant studies have shown that some herbivores feed on phytochemical-containing plants reducing both parasite abundance within hosts [2] and their virulence [3].
Suffering less from parasites is clearly beneficial for infected herbivores. Why then, is this behaviour not fixed in nature? There are, at least, two possible explanations. First, feeding on ‘medicinal’, often toxic, plants may impose costs upon uninfected individuals. Second, parasites may evolve resistance to such chemicals. Whereas the first possibility has been explored, and is taken as evidence for ‘self-medication’ [3], the second hypothesis requires investigation.
A recent study by Palmer-Young et al. [4] fills this gap. This article reports evolution of resistance to two different phytochemicals, alone and in combination, in the trypanosome Crithidia bombi, a bumble bee (Bombus impatiens) parasite. To show this, the authors experimentally evolved a strain of C. bombi in the presence of thymol, eugenol or both simultaneously. These phytochemicals are commonly found in the nectar of several plant species, in particular those of the Lamiaceae, a family containing several aromatic herbs. Prior to selection both phytochemicals reduced C. bombi growth by about 50%. However, C. bombi rapidly evolved resistance in both single and the double phytochemical treatments. Moreover, no cost of resistance was detected when evolved parasites were placed in the ancestral, phytochemical-free environment. Therefore, resistance to phytochemicals would be expected to spread rapidly in natural populations of C. bombi. Clearly, thus, the herbivore strategy of sequestering plant secondary chemical compounds as a defence against their pathogens should fail. The question then is ‘why do they still do it’? Can self-medication work in the longer-term for bumblebees?
Well, yes. The very fact that resistance evolved shows that resistance is not fixed in natural C. bombi populations. This is surprising considering that resistance is not costly. This might be due to a number of reasons. Firstly, there may be costs of resistance that were not detected in this experiment. Second, it may not be possible to evolve universal resistance to the heterogeneity present in the phytochemical environment. Indeed, phytochemical environments are highly varied in time and space and bumblebees will visit different plants presenting different phytochemical cocktails throughout the season. Furthermore, migration of bees from populations exposed to different phytochemicals may prevent the fixation of one resistance type.
Or, it may be self-medication behaviour itself that prevents the evolution of resistance? Indeed, in the same way that infected bees seek cooler temperatures to slow growth of a parasitoid fly [5], they may also seek a more varied diet with diverse phytochemicals to which the parasite cannot evolve, but which reduces parasite growth. Further understanding of arthropod self-medication may thus be instrumental to prevent the observed worldwide decline of pollinators [6]. Furthermore, it may inform on mechanisms that prevent rapid evolution of drug resistance in other systems.
References
[1] Duffey SS. 1980. Sequestration of plant natural products by insects. Annual Review of Entomology 25: 447-477. doi: 10.1146/annurev.en.25.010180.002311
[2] Richardson LL et al. 2015. Secondary metabolites in floral nectar reduce parasite infections in bumblebees. Proceedings of the Royal Society of London B 282: 20142471. doi: 10.1098/rspb.2014.2471
[3] Lefèvre T et al. 2010. Evidence for trans-generational medication in nature. Ecology Letters 13: 1485-93. doi: 10.1111/j.1461-0248.2010.01537.x
[4] Palmer-Young EC, Sadd BM, Adler LS. 2017. Evolution of resistance to single and combined floral phytochemicals by a bumble bee parasite. Journal of Evolutionary Biology 30: 300-312. doi: 10.1111/jeb.13002
[5] Müller CB, Schmid-Hempel P. 1993. Exploitation of cold temperature as defence against parasitoids in bumblebees. Nature 363: 65-67. doi: 10.1038/363065a0
[6] Potts SG et al. 2010. Global pollinator declines: trends, impacts and drivers. Trends in Ecology and Evolution 25: 345-353. doi: 10.1016/j.tree.2010.01.007
| Evolution of resistance to single and combined floral phytochemicals by a bumble bee parasite | Palmer-Young EC, Sadd BM, Adler LS | Repeated exposure to inhibitory compounds can drive the evolution of resistance, which weakens chemical defence against antagonists. Floral phytochemicals in nectar and pollen have antimicrobial properties that can ameliorate infection in pollinat... |  | Evolutionary Ecology | Alison Duncan | | 2016-12-14 16:47:14 | View |
Wolbachia and host intrinsic reproductive barriers contribute additively to post-mating isolation in spider mites
Miguel A. Cruz, Sara Magalhães, Élio Sucena, Flore Zélé
https://doi.org/10.1101/2020.06.29.178699
Speciation in spider mites: disentangling the roles of Wolbachia-induced vs. nuclear mating incompatibilities
Recommended by Jan Engelstaedter based on reviews by Wolfgang Miller and 1 anonymous reviewer
Cytoplasmic incompatibility (CI) is a mating incompatibility that is induced by maternally inherited endosymbionts in many arthropods. These endosymbionts include, most famously, the alpha-proteobacterium Wolbachia pipientis (Yen & Barr 1971; Werren et al. 2008) but also the Bacteroidetes bacterium Cardinium hertigii (Zchori-Fein et al. 2001), a gamma-proteobacterium of the genus Rickettsiella (Rosenwald et al. 2020) and another, as yet undescribed alpha-proteobacterium (Takano et al. 2017). CI manifests as embryonic mortality in crosses between infected males and females that are uninfected or infected with a different strain, whereas embryos develop normally in all other crosses. This phenotype may enable the endosymbionts to spread rapidly within their host population. Exploiting this, CI-inducing Wolbachia are being harnessed to control insect-borne diseases (e.g., O'Neill 2018). Much progress elucidating the genetic basis and developmental mechanism of CI has been made in recent years, but many open questions remain (Shropshire et al. 2020).
Immediately following the discovery and early study of CI in mosquitoes, Laven (1959, 1967) proposed that CI could be an important driver of speciation. Indeed, bi-directional CI can strongly reduce gene flow between two populations due to the elimination of F1 embryos, so that CI can act as a trigger for genetic differentiation in the host (Telschow et al. 2002, 2005). This idea has received much attention, and a potential role for CI in incipient speciation has been demonstrated in several species (e.g., Bordenstein et al. 2001; Jaenike et al. 2006). However, we still don’t know how commonly CI actually triggers speciation, rather than being merely a minor player or secondary phenomenon. The problem is that in addition to CI, postzygotic reproductive isolation can also be caused by host-induced, nuclear incompatibilities. Determining the relative contributions of these two causes of isolation is difficult and has rarely been done.
The study by Cruz et al. (2020) addresses this problem head-on, using a study system of Tetranychus urticae spider mites. These cosmopolitan mites are infected with different strains of Wolbachia. They come in two different colour forms (red and green) that can co-occur sympatrically on the same host plant but exhibit various degrees of reproductive isolation. A complicating factor in spider mites is that they are haplodiploid: unfertilised eggs develop into haploid males and are therefore not affected by any postzygotic incompatibilities, whereas fertilised eggs normally develop into diploid females. In haplodiploids, Wolbachia-induced CI can either kill diploid embryos (as in diplodiploid species), or turn them into haploid males. In their study, Cruz et al. used three different populations (one of the green and two of the red form) and employed a full factorial experiment involving all possible combinations of crosses of Wolbachia infected or uninfected males and females. For each cross, they measured F1 embryonic and juvenile mortality as well as sex ratio, and they also measured F1 fertility and F2 viability. Their results showed that there is strong reduction in hybrid female production caused by Wolbachia-induced CI. However, independent of this and through a different mechanism, there is an even stronger reduction in hybrid production caused by host-associated incompatibilities. In combination with the also observed near-complete sterility of F1 hybrid females and full F2 hybrid breakdown (neither of which is caused by Wolbachia), the results indicate essentially complete reproductive isolation between the green and red forms of T. urticae.
Overall, this is an elegant study with an admirably clean and comprehensive experimental design. It demonstrates that Wolbachia can contribute to reproductive isolation between populations, but that host-induced mechanisms of reproductive isolation predominate in these spider mite populations. Further studies in this exiting system would be useful that also investigate the contribution of pre-zygotic isolation mechanisms such as assortative mating, ascertain whether the results can be generalised to other populations, and – most challengingly – establish the order in which the different mechanisms of reproductive isolation evolved.
References
Bordenstein, S. R., O'Hara, F. P., and Werren, J. H. (2001). Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature, 409(6821), 707-710. doi: https://doi.org/10.1038/35055543
Cruz, M. A., Magalhães, S., Sucena, É., and Zélé, F. (2020) Wolbachia and host intrinsic reproductive barriers contribute additively to post-mating isolation in spider mites. bioRxiv, 2020.06.29.178699, ver. 4 peer-reviewed and recommended by PCI Evolutionary Biology. doi: https://doi.org/10.1101/2020.06.29.178699
Jaenike, J., Dyer, K. A., Cornish, C., and Minhas, M. S. (2006). Asymmetrical reinforcement and Wolbachia infection in Drosophila. PLoS Biol, 4(10), e325. doi: https://doi.org/10.1371/journal.pbio.0040325
Laven, H. (1959). SPECIATION IN MOSQUITOES Speciation by Cytoplasmic Isolation in the Culex Pipiens-Complex. In Cold Spring Harbor Symposia on Quantitative Biology (Vol. 24, pp. 166-173). Cold Spring Harbor Laboratory Press.
Laven, H. (1967). A possible model for speciation by cytoplasmic isolation in the Culex pipiens complex. Bulletin of the World Health Organization, 37(2), 263-266.
O’Neill S.L. (2018) The Use of Wolbachia by the World Mosquito Program to Interrupt Transmission of Aedes aegypti Transmitted Viruses. In: Hilgenfeld R., Vasudevan S. (eds) Dengue and Zika: Control and Antiviral Treatment Strategies. Advances in Experimental Medicine and Biology, vol 1062. Springer, Singapore. doi: https://doi.org/10.1007/978-981-10-8727-1_24
Rosenwald, L.C., Sitvarin, M.I. and White, J.A. (2020). Endosymbiotic Rickettsiella causes cytoplasmic incompatibility in a spider host. doi: https://doi.org/10.1098/rspb.2020.1107
Shropshire, J. D., Leigh, B., and Bordenstein, S. R. (2020). Symbiont-mediated cytoplasmic incompatibility: what have we learned in 50 years?. Elife, 9, e61989. doi: https://doi.org/10.7554/eLife.61989
Takano et al. (2017). Unique clade of alphaproteobacterial endosymbionts induces complete cytoplasmic incompatibility in the coconut beetle. Proceedings of the National Academy of Sciences, 114(23), 6110-6115. doi: https://doi.org/10.1073/pnas.1618094114
Telschow, A., Hammerstein, P., and Werren, J. H. (2002). The effect of Wolbachia on genetic divergence between populations: models with two-way migration. the american naturalist, 160(S4), S54-S66. doi: https://doi.org/10.1086/342153
Telschow, A., Hammerstein, P., and Werren, J. H. (2005). The effect of Wolbachia versus genetic incompatibilities on reinforcement and speciation. Evolution, 59(8), 1607-1619. doi: https://doi.org/10.1111/j.0014-3820.2005.tb01812.x
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nature Reviews Microbiology, 6(10), 741-751. doi: https://doi.org/10.1038/nrmicro1969
Yen, J. H., and Barr, A. R. (1971). New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature, 232(5313), 657-658. doi: https://doi.org/10.1038/232657a0
Zchori-Fein, E., Gottlieb, Y., Kelly, S. E., Brown, J. K., Wilson, J. M., Karr, T. L., and Hunter, M. S. (2001). A newly discovered bacterium associated with parthenogenesis and a change in host selection behavior in parasitoid wasps. Proceedings of the National Academy of Sciences, 98(22), 12555-12560. doi: https://doi.org/10.1073/pnas.221467498
| Wolbachia and host intrinsic reproductive barriers contribute additively to post-mating isolation in spider mites | Miguel A. Cruz, Sara Magalhães, Élio Sucena, Flore Zélé | <p>Wolbachia are widespread maternally-inherited bacteria suggested to play a role in arthropod host speciation through induction of cytoplasmic incompatibility, but this hypothesis remains controversial. Most studies addressing Wolbachia-induced ... |  | Evolutionary Ecology, Hybridization / Introgression, Life History, Reproduction and Sex, Speciation, Species interactions | Jan Engelstaedter | | 2020-07-09 10:18:28 | View |
Multi-gene and lineage comparative assessment of the strength of selection in Hymenoptera
Recommended by Bertanne Visser based on reviews by Michael Lattorff and 1 anonymous reviewer
Genetic variation is the raw material for selection to act upon and the amount of genetic variation present within a population is a pivotal determinant of a population’s evolutionary potential. A large effective population size, i.e., the ideal number of individuals experiencing the same amount of genetic drift and inbreeding as an actual population, Ne (Wright 1931, Crow 1954), thus increases the probability of long-term survival of a population. However, natural populations, as opposed to theoretical ones, rarely adhere to the requirements of an ideal panmictic population (Sjödin et al. 2005). A range of circumstances can reduce Ne, including the structuring of populations (through space and time, as well as age and developmental stages) and inbreeding (Charlesworth 2009). In mammals, species with a larger body mass (as a proxy for lower Ne) were found to have a higher rate of nonsynonymous nucleotide substitutions (that alter the amino acid sequence of a protein), as well as radical amino acid substitutions (altering the physicochemical properties of a protein) (Popadin et al. 2007). In general, low effective population sizes increase the chance of mutation accumulation and drift, while reducing the strength of selection (Sjödin et al. 2005).
In this paper, Weyna and Romiguier (2021) set out to test if parasitism, body size, geographic range, and/or eusociality affect the strength of selection in Hymenoptera. Hymenoptera include the bees, wasps and ants and is an extraordinarily diverse order within the insects. It was recently estimated that Hymenoptera is the most speciose order of the animal kingdom (Forbes et al. 2018). Hymenoptera are further characterized by an impressive radiation of parasitic species, mainly parasitoids, that feed in or on a single host individual to complete their own development (Godfray 1994). All hymenopterans share the same sex determination system: haplo-diploidy, where unfertilized eggs are haploid males and fertilized eggs are diploid females. Compared to other animals, Hymenoptera further contain an impressive number of clades that evolved eusociality (Rehan and Toth 2015), in which societies show a clear division of labor for reproduction (i.e., castes) and cooperative brood care. Hymenopterans thus represent a diverse and interesting group of insects to investigate potential factors affecting strength of selection and Ne.
Using a previously published phylogenomic dataset containing 3256 genes and 169 hymenopteran species (Peters et al. 2017), Weyna and Romiguier (2021) estimated mean genomic dN/dS ratios (nonsynonymous to synonymous substitution rates) for each species and compared these values between parasitic and non-parasitic species, eusocial and solitary species, and in relation to body size, parasitoid-specific traits and geographic range, thought to affect the effective population size and strength of selection. The use of a large number of species, as well as several distinct traits is a clear asset of this study. The authors found no effect of body size, geographic range or parasitism (including a range of parasitoid-specific traits). There was an effect, however, of eusociality where dN/dS increased in three out of four eusocial lineages. Future studies including more independent evolutionary transitions to eusociality can lend further support that eusocial species indeed reduces the efficiency of selection. The most intriguing result was that for solitary and social bees, with high dN/dS ratios and a strong signature of relaxed selection (i.e., the elimination or reduction of a source of selection (Lahti et al. 2009). The authors suggest that the pollen-collecting behaviors of these species can constrain Ne, as pollen availability varies at both a spatial and temporal scale, requiring a large investment in foraging that may in turn limit reproductive output. It would be interesting to see if other pollen feeders, such as certain beetles, flies, butterflies and moths, as well as mites and spiders, experience relaxed selection as a consequence of the trade-off between energy investment in pollen foraging versus fecundity.
References
Charlesworth, B. (2009). Effective population size and patterns of molecular evolution and variation. Nature Reviews Genetics, 10(3), 195-205. doi: https://doi.org/10.1038/nrg2526
Crow, J. F. (1954) Statistics and Mathematics in Biology (eds Kempthorne, O., Bancroft, T. A., Gowen, J. W. & Lush, J. L.) 543–556 (Iowa State Univ. Press, Ames, Iowa)
Forbes, A. A., Bagley, R. K., Beer, M. A., Hippee, A. C., and Widmayer, H. A. (2018). Quantifying the unquantifiable: why Hymenoptera, not Coleoptera, is the most speciose animal order. BMC ecology, 18(1), 1-11. doi: https://doi.org/10.1186/s12898-018-0176-x
Godfray, H. C. J. (1994) Parasitoids: Behavioral and Evolutionary Ecology. Vol. 67, Princeton University Press, 1994. doi: https://doi.org/10.2307/j.ctvs32rmp
Lahti et al. (2009). Relaxed selection in the wild. Trends in ecology & evolution, 24(9), 487-496. doi: https://doi.org/10.1016/j.tree.2009.03.010
Peters et al. (2017). Evolutionary history of the Hymenoptera. Current Biology, 27(7), 1013-1018. doi: https://doi.org/10.1016/j.cub.2017.01.027
Popadin, K., Polishchuk, L. V., Mamirova, L., Knorre, D., and Gunbin, K. (2007). Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proceedings of the National Academy of Sciences, 104(33), 13390-13395. doi: https://doi.org/10.1073/pnas.0701256104
Rehan, S. M., and Toth, A. L. (2015). Climbing the social ladder: the molecular evolution of sociality. Trends in ecology & evolution, 30(7), 426-433. doi: https://doi.org/10.1016/j.tree.2015.05.004
Sjödin, P., Kaj, I., Krone, S., Lascoux, M., and Nordborg, M. (2005). On the meaning and existence of an effective population size. Genetics, 169(2), 1061-1070. doi: https://doi.org/10.1534/genetics.104.026799
Weyna, A., and Romiguier, J. (2021) Relaxation of purifying selection suggests low effective population size in eusocial Hymenoptera and solitary pollinating bees. bioRxiv, 2020.04.14.038893, ver. 5 peer-reviewed and recommended by PCI Evol Biol. doi: https://doi.org/10.1101/2020.04.14.038893
Wright, S. (1931). Evolution in Mendelian populations. Genetics, 16(2), 97-159.
| Relaxation of purifying selection suggests low effective population size in eusocial Hymenoptera and solitary pollinating bees | Arthur Weyna, Jonathan Romiguier | <p>With one of the highest number of parasitic, eusocial and pollinator species among all insect orders, Hymenoptera features a great diversity of lifestyles. At the population genetic level, such life-history strategies are expected to decrease e... |  | Behavior & Social Evolution, Genome Evolution, Life History, Molecular Evolution, Population Genetics / Genomics | Bertanne Visser | | 2020-04-21 17:30:57 | View |
Transgenerational plasticity of inducible defenses: combined effects of grand-parental, parental and current environments
Juliette Tariel; Sandrine Plénet; Emilien Luquet
https://doi.org/10.1101/589945
Transgenerational plasticity through three generations
Recommended by Troy Day based on reviews by Stewart Plaistow and 1 anonymous reviewer
Organisms very often display phenotypic plasticity, whereby the expression of trait (or suite of traits) changes in a consistent way as a function of some environmental variable. Sometimes this plastic response remains labile and so the trait continues to respond to the environment throughout an organism’s life, but there are also many examples in which environmental conditions during a critical developmental window irreversibly set the stage for how a trait will be expressed later in life.
Traditionally, most studies of phenotypic plasticity have considered how an organism’s phenotype is altered by the environment that it experiences (called within-generation plasticity) but there is growing interest in how an organism’s phenotype is altered by the environment experienced by its ancestors (called transgenerational plasticity) [1]. In the simplest cases an organism’s phenotype might be affected by the environmental conditions experienced by its parents. There are several examples of this phenomenon as well, including interesting cases where predator cues experiences by an organism’s parents dictate the extent to which it displays a defensive phenotype.
Tariel et al. [2] present a study that takes these ideas to the next logical step and examines transgenerational plasticity through three generations. They used a well-studied system of snails (Physa acuta) that display inducible defences in response to predator (crayfish) cues. The authors exposed three generations of snails to one of two treatments: the presence or absence of predator cues, and then examined a suite of behavioural and morphological traits associated with predator defence. This allowed them to determine if and how offspring, parental, and grandparental environment influence offspring phenotype.
Interestingly, their results do show that transgenerational plasticity can act across multiple generations. The patterns found were complex though and it is difficult at this stage to assess how likely it is that these responses are adaptive. For example, a behavioural trait appears to respond to grandparental but not parental environment, shell thickness responds to both, and snail weight and a composite index of morphology respond to neither. Exactly what this means in terms of an offspring’s fitness, however, is unclear. It is also not immediately clear from the study how predictive a grandparent’s environment is of the conditions likely to be faced by an individual. Further work will be needed on these issues to better interpret what this transgenerational plasticity means and to assess if it might be an evolved response to cope with varying predation pressure. It would also be useful to delve more deeply into the developmental mechanisms throughout which this plasticity occurs. Irrespective of these issues, however, the study does reveal that transgenerational plasticity across multiple generations can indeed occur and so cannot be ignored as a source of phenotypic variation.
References
[1] West-Eberhard, M. J. (2003). Developmental plasticity and evolution. Oxford University Press.
[2] Tariel, J., Plenet, S., and Luquet, E. (2019). Transgenerational plasticity of inducible defenses: combined effects of grand-parental, parental and current environments. bioRxiv, 589945, ver. 3, peer-reviewed and recommended by Peer Community in Evolutionary Biology. doi: 10.1101/589945
| Transgenerational plasticity of inducible defenses: combined effects of grand-parental, parental and current environments | Juliette Tariel; Sandrine Plénet; Emilien Luquet | <p>While an increasing number of studies highlights that parental environment shapes offspring phenotype (transgenerational plasticity TGP), TGP beyond the parental generation has received less attention. Studies suggest that TGP impacts populatio... |  | Adaptation, Evolutionary Ecology, Non Genetic Inheritance, Phenotypic Plasticity | Troy Day | | 2019-03-29 09:31:53 | View |
Field evidence for manipulation of mosquito host selection by the human malaria parasite, Plasmodium falciparum
Amelie Vantaux, Franck Yao, Domonbabele FdS Hien, Edwige Guissou, Bienvenue K Yameogo, Louis-Clement Gouagna, Didier Fontenille, Francois Renaud, Frederic Simard, Carlo Constantini, Frederic Thomas, Karine Mouline, Benjamin Roche, Anna Cohuet, Kounbobr R Dabire, Thierry Lefevre
https://doi.org/10.1101/207183
Malaria host manipulation increases probability of mosquitoes feeding on humans
Recommended by Alison Duncan based on reviews by Olivier Restif, Ricardo S. Ramiro and 1 anonymous reviewer
Parasites can manipulate their host’s behaviour to ensure their own transmission. These manipulated behaviours may be outside the range of ordinary host activities [1], or alter the crucial timing and/or location of a host’s regular activity. Vantaux et al show that the latter is true for the human malaria parasite, Plasmodium falciparum [2]. They demonstrate that three species of Anopheles mosquito were 24% more likely to choose human hosts, rather than other vertebrates, for their blood feed when they harboured transmissible stages (sporozoites) compared to when they were uninfected, or infected with non-transmissible malaria parasites [2]. Host choice is crucial for the malaria parasite Plasmodium falciparum to complete its life-cycle, as their host range is much narrower than the mosquito’s for feeding; P. falciparum can only develop in hominids, or closely related apes [3].
The study only shows this stage-dependent parasite manipulation retrospectively (by identifying host type and parasite stage in mosquitoes after their blood feed [2]). There was no difference in the preferences of infectious (with sporozoites) or un-infectious (infected without sporozoites, or uninfected) mosquitoes between human versus cow hosts in a choice test [2]. This suggests that the final decision about whether to feed occurs when the mosquito is in close range of the host.
This, coupled with previous findings, shows that vector manipulation is a fine-tuned business, that can act at multiple stages of the parasite life-cycle and on many behaviours [4]. Indeed, mosquitoes with non-transmissible Plasmodium stages (oocysts) are more reluctant to feed than sporozoite-infected mosquitoes [5] as vectors can be killed by their host whilst feeding, doing so before they are ready to transmit is risky for the malaria parasite. Thus, it seems that Plasmodium is, to some extent, master of its vector; commanding it not to feed when it cannot be transmitted, to feed when it is ready to be transmitted and to feed on the right type of host. What does this mean for our understanding of malaria transmission and epidemics?
Vantaux et al use a mathematical model, parameterised using data from this experiment, to highlight the consequences of this 24% increase in feeding on humans for P. falciparum transmission. They show that this increase raises the number of infectious bites humans receive from 4 (if sporozoite-infected mosquitoes had the same probability as uninfected mosquitoes) to 14 (an increase in 250%), for mosquitoes with a 15-day life-span, at ratios of 1:1 mosquitoes to humans. Longer mosquito life-spans and higher ratios of mosquitoes to humans further increases the number of infectious bites.
These results [2] have important implications for epidemiological forecasting and disease management. Public health strategies could focus on possible ways to trap sporozoite-infected mosquitoes, mimicking cues they use to locate their human hosts, or identify the behaviour of mosquitoes harbouring non-yet infectious Plasmodium, and trap them before they bite. Moreover, the results of the model show that failing to take into account the preference for humans of sporozoite-infected mosquitoes could underestimate the size of pending epidemics.
An important question previously raised is whether Plasmodium-induced alteration in host behaviour really is manipulation, or just a side-effect of being infected [4,5]. The fact that Vantaux et al show that these altered feeding behaviours increases the likelihood of transmission, in that a sporozoite-infected mosquito is more likely to feed on a human, strongly suggests that it is adaptive for the parasite [2]. Ultimately, to show that it is manipulation would require the identification of molecular factors released by Plasmodium that are responsible for physiological changes in the mosquito [6].
References
[1] Thomas, F., Schmidt-Rhaesa, A., Martin, G., Manu, C., Durand, P., & Renaud, F. (2002). Do hairworms (Nematomorpha) manipulate the water seeking behaviour of their terrestrial hosts? Journal of Evolutionary Biology, 15(3), 356–361. doi: 10.1046/j.1420-9101.2002.00410.x
[2] Vantaux, A., Yao, F., Hien, D. F., Guissou, E., Yameogo, B. K., Gouagna, L.-C., … Lefevre, T. (2018). Field evidence for manipulation of mosquito host selection by the human malaria parasite, Plasmodium falciparum. BioRxiv, 207183 ver 6. doi: 10.1101/207183
[3] Prugnolle, F., Durand, P., Ollomo, B., Duval, L., Ariey, F., Arnathau, C., … Renaud, F. (2011). A Fresh Look at the Origin of Plasmodium falciparum, the Most Malignant Malaria Agent. PLOS Pathogens, 7(2), e1001283. doi: 10.1371/journal.ppat.1001283
[4] Cator, L. J., Lynch, P. A., Read, A. F., & Thomas, M. B. (2012). Do malaria parasites manipulate mosquitoes? Trends in Parasitology, 28(11), 466–470. doi: 10.1016/j.pt.2012.08.004
[5] Cator, L. J., George, J., Blanford, S., Murdock, C. C., Baker, T. C., Read, A. F., & Thomas, M. B. (2013). “Manipulation” without the parasite: altered feeding behaviour of mosquitoes is not dependent on infection with malaria parasites. Proceedings. Biological Sciences, 280(1763), 20130711. doi: 10.1098/rspb.2013.0711
[6] Herbison, R., Lagrue, C., & Poulin, R. (2018). The missing link in parasite manipulation of host behaviour. Parasites & Vectors, 11. doi: 10.1186/s13071-018-2805-9
| Field evidence for manipulation of mosquito host selection by the human malaria parasite, Plasmodium falciparum | Amelie Vantaux, Franck Yao, Domonbabele FdS Hien, Edwige Guissou, Bienvenue K Yameogo, Louis-Clement Gouagna, Didier Fontenille, Francois Renaud, Frederic Simard, Carlo Constantini, Frederic Thomas, Karine Mouline, Benjamin Roche, Anna Cohuet, Kou... | <p>Whether the malaria parasite *Plasmodium falciparum* can manipulate mosquito host choice in ways that enhance parasite transmission toward human is unknown. We assessed the influence of *P. falciparum* on the blood-feeding behaviour of three of... |  | Evolutionary Ecology | Alison Duncan | | 2018-02-28 09:12:14 | View |
Is adaptation limited by mutation? A timescale-dependent effect of genetic diversity on the adaptive substitution rate in animals
Marjolaine Rousselle, Paul Simion, Marie-Ka Tilak, Emeric Figuet, Benoit Nabholz, Nicolas Galtier
https://doi.org/10.1101/643619
To tinker, evolution needs a supply of spare parts
Recommended by Georgii Bazykin based on reviews by Konstantin Popadin, David Enard and 1 anonymous reviewer
Is evolution adaptive? Not if there is no variation for natural selection to work with. Theory predicts that how fast a population can adapt to a new environment can be limited by the supply of new mutations coming into it. This supply, in turn, depends on two things: how often mutations occur and in how many individuals. If there are few mutations, or few individuals in whom they can originate, individuals will be mostly identical in their DNA, and natural selection will be impotent.
This theoretical prediction has been hard to test. The rate at which new mutations arise in a population can be manipulated experimentally, and some work has shown that the fitness of a population increases more rapidly if more new mutations appear per generation, lending support to the mutation-limitation hypothesis [1]. However, the question remains whether this limitation has played a role in the history of life over the evolutionary timescale. Maybe all natural populations are so large, the mutation rate so high, and/or the environment changes so slowly, that any novel variant required for adaptation is already there when selection starts to act? Some recent work does suggest that when strong selection begins to favor a certain phenotype, multiple distinct genetic variants producing this phenotype spread; this is what has happened, for instance, at the origin of insecticide resistance in wild populations of Drosophila melanogaster [2] or lactose persistence in humans [3]. In many other cases, though, adaptations seem to originate through a single mutation event, suggesting that the time needed for this mutation to arise may be important.
To complicate things, adaptation is hard to quantify. It leaves a trace in differences between individuals of the same species as well as of different species. However, this trace is often masked or confounded by other processes, including natural selection disfavoring newly arising deleterious variants, interference from selection acting at linked sites, and changes in population size. In 1991, McDonald and Kreitman [4] have come up with a method to infer the rate of adaptation in the presence of strong negative selection, and later work has developed upon it to control for some of the other confounders. Still, the method is data-intensive, and previous attempts to employ it to compare the rates of adaptation between species have yielded somewhat contradictory results.
The new paper by Rousselle et al. recommended by PCI Evol Biol [5] fills this gap. The authors use published data as well as their own newly generated dataset to analyze, in a McDonald and Kreitman-like framework, both closely and distantly related species. Importantly, these comparisons cover species with very different polymorphism levels, spanning two orders of magnitude of difference levels.
So is adaptation in fact limited by supply of new mutations? The answer is, it depends. It does indeed seem that the species with a lower level of polymorphism adapt at a lower rate, consistent with the mutation-limitation hypothesis. However, this only is true for those groups of species in which the variability is low. Therefore, if a population is very small or the mutation rate very low, there may be in fact not enough mutations to secure its need to adapt.
In more polymorphic species, and in comparisons of distant species, the data hint instead at the opposite relationship: the rate of adaptations declines with variability. This is consistent with a different explanation: when a population is small, it needs to adapt more frequently, repairing the weakly deleterious mutations that can’t be prevented by selection under small population sizes.
There are quite a few problems small populations have to deal with. Some of them are ecological: e.g., small numbers make populations more vulnerable to stochastic fluctuations in size or sex ratio. Others, however, are genetic. Small populations are prone to inbreeding depression and have an increased rate of genetic drift, leading to spread of deleterious alleles. Indeed, selection against deleterious mutations is less efficient when populations are small, and less numerable species accumulate more of such mutations over the course of evolution [6]. The work by Rouselle et al. [5] suggests that small populations also face an additional burden: a reduced ability to adapt.
Has the rate of adaptation in our own species also been limited by our deficit of diversity? The data hints at this. Homo sapiens, as well as the two other studied extinct representatives of the genus Homo, Neanderthals and Denisovans, belong to the domain of relatively low polymorphism levels, where an increase in polymorphism matters for the rate at which adaptive substitutions accumulate. Perhaps, if our ancestors were more numerous or more mutable, they would have been able to get themselves out of trouble, and there would be multiple human species still alive rather than just one.
References
[1] G, J. A., Visser, M. de, Zeyl, C. W., Gerrish, P. J., Blanchard, J. L., and Lenski, R. E. (1999). Diminishing Returns from Mutation Supply Rate in Asexual Populations. Science, 283(5400), 404–406. doi: 10.1126/science.283.5400.404
[2] Karasov, T., Messer, P. W., and Petrov, D. A. (2010). Evidence that Adaptation in Drosophila Is Not Limited by Mutation at Single Sites. PLOS Genetics, 6(6), e1000924. doi: 10.1371/journal.pgen.1000924
[3] Jones, B. L., Raga, T. O., Liebert, A., Zmarz, P., Bekele, E., Danielsen, E. T., Olsen, A. K., Bradman, N., Troelsen, J. T., and Swallow, D. M. (2013). Diversity of Lactase Persistence Alleles in Ethiopia: Signature of a Soft Selective Sweep. The American Journal of Human Genetics, 93(3), 538–544. doi: 10.1016/j.ajhg.2013.07.008
[4] McDonald, J. H., and Kreitman, M. (1991). Adaptive protein evolution at the Adh locus in Drosophila. Nature, 351(6328), 652–654. doi: 10.1038/351652a0
[5] Rousselle, M., Simion, P., Tilak, M. K., Figuet, E., Nabholz, B., and Galtier, N. (2019). Is adaptation limited by mutation? A timescale-dependent effect of genetic diversity on the adaptive substitution rate in animals. BioRxiv, 643619, ver 4 peer-reviewed and recommended by Peer Community In Evolutionary Biology. doi: 10.1101/643619
[6] Popadin, K., Polishchuk, L. V., Mamirova, L., Knorre, D., and Gunbin, K. (2007). Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proceedings of the National Academy of Sciences, 104(33), 13390–13395. doi: 10.1073/pnas.0701256104
| Is adaptation limited by mutation? A timescale-dependent effect of genetic diversity on the adaptive substitution rate in animals | Marjolaine Rousselle, Paul Simion, Marie-Ka Tilak, Emeric Figuet, Benoit Nabholz, Nicolas Galtier | <p>Whether adaptation is limited by the beneficial mutation supply is a long-standing question of evolutionary genetics, which is more generally related to the determination of the adaptive substitution rate and its relationship with the effective... |  | Adaptation, Evolutionary Theory, Genome Evolution, Molecular Evolution, Population Genetics / Genomics | Georgii Bazykin | | 2019-05-21 09:49:16 | View |
A general model of fitness effects following hybridisation
Recommended by Matthew Hartfield based on reviews by Luis-Miguel Chevin and Juan Li
Studying the effects of speciation, hybridisation, and evolutionary outcomes following reproduction from divergent populations is a major research area in evolutionary genetics [1]. There are two phenomena that have been the focus of contemporary research. First, a classic concept is the formation of ‘Bateson-Dobzhansky-Muller’ incompatibilities (BDMi) [2–4] that negatively affect hybrid fitness. Here, two diverging populations accumulate mutations over time that are unique to that subpopulation. If they subsequently meet, then these mutations might negatively interact, leading to a loss in fitness or even a complete lack of reproduction. BDMi formation can be complex, involving multiple genes and the fitness changes can depend on the direction of introgression [5]. Second, such secondary contact can instead lead to heterosis, where offspring are fitter than their parental progenitors [6].
Understanding which outcomes are likely to arise require one to know the potential fitness effects of mutations underlying reproductive isolation, to determine whether they are likely to reduce or enhance fitness when hybrids are formed. This is far from an easy task, as it requires one to track mutations at several loci, along with their effects, across a fitness landscape.
The work of De Sanctis et al. [7] neatly fills in this knowledge gap, by creating a general mathematical framework for describing the consequences of a cross from two divergent populations. The derivations are based on Fisher’s Geometric Model, which is widely used to quantify selection acting on a general fitness landscape that is affected by several biological traits [8,9], and has previously been used in theoretical studies of hybridisation [10–12]. By doing so, they are able to decompose how divergence at multiple loci affects offspring fitness through both additive and dominance effects.
A key result arising from their analyses is demonstrating how offspring fitness can be captured by two main functions. The first one is the ‘net effect of evolutionary change’ that, broadly defined, measures how phenotypically divergent two populations are. The second is the ‘total amount of evolutionary change’, which reflects how many mutations contribute to divergence and the effect sizes captured by each of them. The authors illustrate these measurements using simulations covering different scenarios, demonstrating how different parental states can lead to similar fitness outcomes. They also propose experimental methods to measure the underlying mutational effects.
This study neatly demonstrates how complex genetic phenomena underlying hybridisation can be captured using fairly simple mathematical formulae. This powerful approach will thus open the door for future research to investigate hybridisation in more detail, whether it is by expanding on these theoretical models or using the elegant outcomes to quantify fitness effects in experiments.
References
1. Coyne JA, Orr HA. Speciation. Sunderland, Mass: Sinauer Associates; 2004.
2. Bateson W, Seward A. Darwin and modern science. Heredity and variation in modern lights. 1909;85: 101. https://doi.org/10.1017/CBO9780511693953.007
3. Dobzhansky T. Genetics and the Origin of Species. Columbia university press; 1937.
4. Muller HJ. Isolating mechanisms, evolution and temperature. Biol Symp. 1942;6: 71-125.
5. Fraïsse C, Elderfield JAD, Welch JJ. The genetics of speciation: are complex incompatibilities easier to evolve? J Evol Biol. 2014;27: 688-699. https://doi.org/10.1111/jeb.12339
6. Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA. Heterosis. The Plant Cell. 2010;22: 2105-2112. https://doi.org/10.1105/tpc.110.076133
7. De Sanctis B, Schneemann H, Welch JJ. How does the mode of evolutionary divergence affect reproductive isolation? bioRxiv. 2022. 2022.03.08.483443 version 4. https://doi.org/10.1101/2022.03.08.483443
8. Fisher RA. The genetical theory of natural selection. Oxford: The Clarendon Press; 1930. https://doi.org/10.5962/bhl.title.27468
9. Tenaillon O. The Utility of Fisher's Geometric Model in Evolutionary Genetics. Annu Rev Ecol Evol Syst. 2014;45: 179-201. https://doi.org/10.1146/annurev-ecolsys-120213-091846
10. Barton NH. The role of hybridization in evolution. Molecular Ecology. 2001;10: 551-568. https://doi.org/10.1046/j.1365-294x.2001.01216.x
11. Chevin L-M, Decorzent G, Lenormand T. Niche Dimensionality and The Genetics of Ecological Speciation. Evolution. 2014;68: 1244-1256. https://doi.org/10.1111/evo.12346
12. Fraïsse C, Gunnarsson PA, Roze D, Bierne N, Welch JJ. The genetics of speciation: Insights from Fisher's geometric model. Evolution. 2016;70: 1450-1464. https://doi.org/10.1111/evo.12968
| How does the mode of evolutionary divergence affect reproductive isolation? | Bianca De Sanctis, Hilde Schneemann, John J. Welch | <p>When divergent populations interbreed, the outcome will be affected by the genomic and phenotypic differences that they have accumulated. In this way, the mode of evolutionary divergence between populations may have predictable consequences for... | 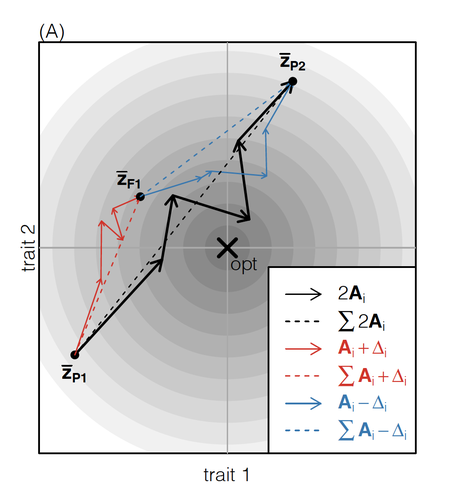 | Adaptation, Evolutionary Theory, Hybridization / Introgression, Population Genetics / Genomics, Speciation | Matthew Hartfield | | 2022-03-30 14:55:46 | View |
Another step towards grasping the complexity of the environmental response of traits
Recommended by Benoit Pujol based on reviews by 2 anonymous reviewers
One can only hope that one day, we will be able to evaluate how the ecological complexity surrounding natural populations affects their ability to adapt. This is more like a long term quest than a simple scientific aim. Many steps are heading in the right direction. This paper by Monroe and colleagues (2021) is one of them.
Many ecological and genetic mechanisms shape the evolutionary potential of phenotypic trait variation and many of them involve environmental heterogeneity (Pujol et al 2018). To date, we cannot look into these ecological and genetic mechanisms without oversimplifying their effects. We often look into trait variation one trait at a time albeit the variation of multiple phenotypic traits is often linked at the genetic or environmental level. As a consequence, we put our conclusions at risk by not accounting for the reciprocal impacts of trait changes upon each other (Teplitsky et al 2014). We also usually restrict the study of a continuous gradient of environmental conditions to a few conditions because it would otherwise be impossible to model its environmental effect. As a consequence, we miss the full picture of the continuous often nonlinear phenotypic plastic response. Whether the trait undergo threshold effect changes thereby remains obscured to us. Collectively, these issues impede our ability to understand how selection shapes the ecological strategy of organisms under variable environments.
In this paper, Monroe and colleagues (2021) propose an original approach that raised to these two challenges. They analysed phenotypic plastic changes in response to a continuous environment in a multidimensional trait space, namely the response of Brachypodium plant developmental and physiological traits to a continuous gradient of soil moisture. They used dry down experimental treatments to produce the continuous soil moisture gradient and compared the plant capacity to use water between annual B. distachyon and perennial B. sylvaticum. Their results revealed the best mathematical functions that model the nonlinear curvature of the continuous plastic response of Brachypodium plants. This work reinforces our view that nonlinear plastic responses can result in greater or lesser trait values at any stage of the environmental gradient that were unexpected on the basis of linear predictors (Gienapp and Brommer 2014). Their findings also imply that different threshold responses characterize different genotypes. These could otherwise have been missed by a classical approach. By shedding light on unforeseen interactions between traits that make their correlation vary along the nonlinear response, they were able to describe more accurately Brachypodium ecological strategies and the changes in evolutionary constraints along the soil moisture gradient.
Their empirical approach allows to test what environmental conditions maximises the opportunity for selection to shape trait variation. For example, it revealed unforeseen divergence in potentially adaptive mechanisms or life history strategies – and not just trait values – between annual and perennial species of Brachypodium. Behind every environmental variation of the constraints to the future evolutionary change of multiple traits, we can expect that the evolutionary history of the populations shaped their trait genetic correlations. Investigating the nonlinear signature of adaptive evolution across continuous environments will get us into uncharted territory.
Our ability to predict the adaptive potential of species is limited. With their approach of continuous environmental gradients beyond linearity, Monroe and collaborators (2021) improve our understanding of plant phenotypic responses and open a brand new range of exciting developments. As they mention: "the opportunity for scaling up" their approach is big. To illustrate this prospect, I can easily think of an example: the quantitative genetic random regression model. This model allows to use any degree of genetic relatedness in a wild population to estimate the genetic variation of phenotypic plastic reaction norms (Nussey et al 2007, Pujol and Galaud 2013). However, in this approach, only a few modalities of the environmental gradient are used to model nonlinear phenotypic plastic responses. From there, it is rather intuitive. Combining the best of these two approaches (continuity of genetic relatedness in the wild & continuity of environmental gradient in experiments) could open ground breaking new perspectives in research.
References
Gienapp P. & J.E. Brommer. 2014. Evolutionary dynamics in response to climate change. In: Charmentier A, Garant D, Kruuk LEB, editors. Quantitative genetics in the wild. Oxford: Oxford University Press, Oxford. pp. 254–273. doi: https://doi.org/10.1093/acprof:oso/9780199674237.003.0015
Monroe, J. G., Cai, H., and Des Marais, D. L. (2020). Trait plasticity and covariance along a continuous soil moisture gradient. bioRxiv, 2020.02.17.952853, ver. 5 peer-reviewed and recommended by PCI Evol Biol. doi: https://doi.org/10.1101/2020.02.17.952853
Pujol et al. (2018). The missing response to selection in the wild. Trends in ecology & evolution, 33(5), 337-346. doi: https://doi.org/10.1016/j.tree.2018.02.007
Pujol, B., and Galaud, J. P. (2013). A practical guide to quantifying the effect of genes underlying adaptation in a mixed genomics and evolutionary ecology approach. Botany Letters, 160(3-4), 197-204. doi: https://doi.org/10.1080/12538078.2013.799045
Nussey, D. H., Wilson, A. J., and Brommer, J. E. (2007). The evolutionary ecology of individual phenotypic plasticity in wild populations. Journal of evolutionary biology, 20(3), 831-844. doi: https://doi.org/10.1111/j.1420-9101.2007.01300.x
Teplitsky et al. (2014). Assessing multivariate constraints to evolution across ten long-term avian studies. PLoS One, 9(3), e90444. doi: https://doi.org/10.1371/journal.pone.0090444
| Trait plasticity and covariance along a continuous soil moisture gradient | J Grey Monroe, Haoran Cai, David L Des Marais | <p>Water availability is perhaps the greatest environmental determinant of plant yield and fitness. However, our understanding of plant-water relations is limited because it is primarily informed by experiments considering soil moisture variabilit... |  | Phenotypic Plasticity | Benoit Pujol | | 2020-02-20 16:34:40 | View |
The quasi-universality of nestedness in the structure of quantitative plant-parasite interactions
Moury Benoît, Audergon Jean-Marc, Baudracco-Arnas Sylvie, Ben Krima Safa, Bertrand François, Boissot Nathalie, Buisson Mireille, Caffier Valérie, Cantet Mélissa, Chanéac Sylvia, Constant Carole, Delmotte François, Dogimont Catherine, Doumayrou Juliette, Fabre Frédéric, Fournet Sylvain, Grimault Valérie, Jaunet Thierry, Justafré Isabelle, Lefebvre Véronique, Losdat Denis, Marcel Thierry C., Montarry Josselin, Morris Cindy E., Omrani Mariem, Paineau Manon, Perrot Sophie, Pilet-Nayel Marie-Laure, R...
https://doi.org/10.1101/2021.03.03.433745
Nestedness and modularity in plant-parasite infection networks
Recommended by Santiago Elena based on reviews by Rubén González and 2 anonymous reviewers based on reviews by Rubén González and 2 anonymous reviewers
In a landmark paper, Flores et al. (2011) showed that the interactions between bacteria and their viruses could be nicely described using a bipartite infection networks. Two quantitative properties of these networks were of particular interest, namely modularity and nestedness. Modularity emerges when groups of host species (or genotypes) shared groups of viruses. Nestedness provided a view of the degree of specialization of both partners: high nestedness suggests that hosts differ in their susceptibility to infection, with some highly susceptible host genotypes selecting for very specialized viruses while strongly resistant host genotypes select for generalist viruses. Translated to the plant pathology parlance, this extreme case would be equivalent to a gene-for-gene infection model (Flor 1956): new mutations confer hosts with resistance to recently evolved viruses while maintaining resistance to past viruses. Likewise, virus mutations for expanding host range evolve without losing the ability to infect ancestral host genotypes. By contrast, a non-nested network would represent a matching-allele infection model (Frank 2000) in which each interacting organism evolves by losing its capacity to resist/infect its ancestral partners, resembling a Red Queen dynamic. Obviously, the reality is more complex and may lie anywhere between these two extreme situations.
Recently, Valverde et al. (2020) developed a model to explain the emergence of nestedness and modularity in plant-virus infection networks across diverse habitats. They found that local modularity could coexist with global nestedness and that intraspecific competition was the main driver of the evolution of ecosystems in a continuum between nested-modular and nested networks. These predictions were tested with field data showing the association between plant host species and different viruses in different agroecosystems (Valverde et al. 2020). The effect of interspecific competition in the structure of empirical plant host-virus infection networks was also tested by McLeish et al. (2019). Besides data from agroecosystems, evolution experiments have also shown the pervasive emergence of nestedness during the diversification of independently-evolved lineages of potyviruses in Arabidopsis thaliana genotypes that differ in their susceptibility to infection (Hillung et al. 2014; González et al. 2019; Navarro et al. 2020).
In their study, Moury et al. (2021) have expanded all these previous observations to a diverse set of pathosystems that range from viruses, bacteria, oomycetes, fungi, nematodes to insects. While modularity was barely seen in only a few of the systems, nestedness was a common trend (observed in ~94% of all systems). This nestedness, as seen in previous studies and as predicted by theory, emerged as a consequence of the existence of generalist and specialist strains of the parasites that differed in their capacity to infect more or less resistant plant genotypes.
As pointed out by Moury et al. (2021) in their conclusions, the ubiquity of nestedness in plant-parasite infection matrices has strong implications for the evolution and management of infectious diseases.
References
Flor, H. H. (1956). The complementary genic systems in flax and flax rust. In Advances in genetics, 8, 29-54. https://doi.org/10.1016/S0065-2660(08)60498-8
Flores, C. O., Meyer, J. R., Valverde, S., Farr, L., and Weitz, J. S. (2011). Statistical structure of host–phage interactions. Proceedings of the National Academy of Sciences, 108, E288-E297. https://doi.org/10.1073/pnas.1101595108
Frank, S. A. (2000). Specific and non-specific defense against parasitic attack. Journal of Theoretical Biology, 202, 283-304. https://doi.org/10.1006/jtbi.1999.1054
González, R., Butković, A., and Elena, S. F. (2019). Role of host genetic diversity for susceptibility-to-infection in the evolution of virulence of a plant virus. Virus evolution, 5(2), vez024. https://doi.org/10.1093/ve/vez052
Hillung, J., Cuevas, J. M., Valverde, S., and Elena, S. F. (2014). Experimental evolution of an emerging plant virus in host genotypes that differ in their susceptibility to infection. Evolution, 68, 2467-2480. https://doi.org/10.1111/evo.12458
McLeish, M., Sacristán, S., Fraile, A., and García-Arenal, F. (2019). Coinfection organizes epidemiological networks of viruses and hosts and reveals hubs of transmission. Phytopathology, 109, 1003-1010. https://doi.org/10.1094/PHYTO-08-18-0293-R
Moury B, Audergon J-M, Baudracco-Arnas S, Krima SB, Bertrand F, Boissot N, Buisson M, Caffier V, Cantet M, Chanéac S, Constant C, Delmotte F, Dogimont C, Doumayrou J, Fabre F, Fournet S, Grimault V, Jaunet T, Justafré I, Lefebvre V, Losdat D, Marcel TC, Montarry J, Morris CE, Omrani M, Paineau M, Perrot S, Pilet-Nayel M-L and Ruellan Y (2021) The quasi-universality of nestedness in the structure of quantitative plant-parasite interactions. bioRxiv, 2021.03.03.433745, ver. 4 recommended and peer-reviewed by PCI Evolutionary Biology. https://doi.org/10.1101/2021.03.03.433745
Navarro, R., Ambros, S., Martinez, F., Wu, B., Carrasco, J. L., and Elena, S. F. (2020). Defects in plant immunity modulate the rates and patterns of RNA virus evolution. bioRxiv. doi: https://doi.org/10.1101/2020.10.13.337402
Valverde, S., Vidiella, B., Montañez, R., Fraile, A., Sacristán, S., and García-Arenal, F. (2020). Coexistence of nestedness and modularity in host–pathogen infection networks. Nature ecology & evolution, 4, 568-577. https://doi.org/10.1038/s41559-020-1130-9
| The quasi-universality of nestedness in the structure of quantitative plant-parasite interactions | Moury Benoît, Audergon Jean-Marc, Baudracco-Arnas Sylvie, Ben Krima Safa, Bertrand François, Boissot Nathalie, Buisson Mireille, Caffier Valérie, Cantet Mélissa, Chanéac Sylvia, Constant Carole, Delmotte François, Dogimont Catherine, Doumayrou Jul... | <p>Understanding the relationships between host range and pathogenicity for parasites, and between the efficiency and scope of immunity for hosts are essential to implement efficient disease control strategies. In the case of plant parasites, most... | 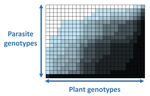 | Bioinformatics & Computational Biology, Evolutionary Dynamics, Species interactions | Santiago Elena | | 2021-03-04 21:23:08 | View |



















 based on reviews by Rubén González and 2 anonymous reviewers
based on reviews by Rubén González and 2 anonymous reviewers











