Experimental evolution of virulence and associated traits in a Drosophila melanogaster – Wolbachia symbiosis
David Monnin, Natacha Kremer, Caroline Michaud, Manon Villa, Hélène Henri, Emmanuel Desouhant, Fabrice Vavre
https://doi.org/10.1101/2020.04.26.062265
Temperature effects on virulence evolution of wMelPop Wolbachia in Drosophila melanogaster
Recommended by Ellen Decaestecker based on reviews by Shira Houwenhuyse and 3 anonymous reviewers
Monnin et al. [1] here studied how Drosophila populations are affected when exposed to a high virulent endosymbiotic wMelPop Wolbachia strain and why virulent vertically transmitting endosymbionts persist in nature. This virulent wMelPop strain has been described to be a blocker of dengue and other arboviral infections in arthropod vector species, such as Aedes aegypti. Whereas it can thus function as a mutualistic symbiont, it here acts as an antagonist along the mutualism-antagonism continuum symbionts operate. The wMelPop strain is not a natural occurring strain in Drosophila melanogaster and thus the start of this experiment can be seen as a novel host-pathogen association. Through experimental evolution of 17 generations, the authors studied how high temperature affects wMelPop Wolbachia virulence and Drosophila melanogaster survival. The authors used Drosophila strains that were selected for late reproduction, given that this should favor evolution to a lower virulence. Assumptions for this hypothesis are not given in the manuscript here, but it can indeed be assumed that energy that is assimilated to symbiont tolerance instead of reproduction may lead to reduced virulence evolution. This has equally been suggested by Reyserhove et al. [2] in a dynamics energy budget model tailored to Daphnia magna virulence evolution upon a viral infection causing White fat Cell disease, reconstructing changing environments through time.
Contrary to their expectations for vertically transmitting symbionts, the authors did not find a reduction in wMelPop Wolbachia virulence during the course of the experimental evolution experiment under high temperature. Important is what this learns for virulence evolution, also for currently horizontal transmitting disease epidemics (such as COVID-19). It mainly reflects that evolution of virulence for new host-pathogen associations is difficult to predict and that it may take multiple generations before optimal levels of virulence are reached [3,4]. These optimal levels of virulence will depend on trade-offs with other life history traits of the symbiont, but also on host demography, host heterogeneity, amongst others [5,6]. Multiple microbial interactions may affect the outcome of virulence evolution [7]. Given that no germ-free individuals were used, it can be expected that other components of the Drosophila microbiome may have played a role in the virulence evolution. In most cases, microbiota have been described as defensive or protective for virulent symbionts [8], but they may also have stimulated the high levels of virulence. Especially, given that upon higher temperatures, Wolbachia growth may have been increased, host metabolic demands increased [9], host immune responses affected and microbial communities changed [10]. This may have resulted in increased competitive interactions to retrieve host resources, sustaining high virulence levels of the symbiont.
A nice asset of this study is that the phenotypic results obtained in the experimental evolution set-up were linked with wMelPop density measurement and octomom copy number quantifications. Octomom is a specific 8-n genes region of the Wolbachia genome responsible for wMelPop virulence, so there is a link between the phenotypic and molecular functions of the involved symbiont. The authors found that density, octomom copy number and virulence were correlated to each other. An important note the authors address in their discussion is that, to exclude the possibility that octomom copy number has an effect on density, and density on virulence, the effect of these variables should be assessed independently of temperature and age. The obtained results are a valuable contribution to the ongoing debate on the relationship between wMelPop octomom copy number, density and virulence.
References
[1] Monnin, D., Kremer, N., Michaud, C., Villa, M., Henri, H., Desouhant, E. and Vavre, F. (2020) Experimental evolution of virulence and associated traits in a Drosophila melanogaster – Wolbachia symbiosis. bioRxiv, 2020.04.26.062265, ver. 4 peer-reviewed and recommended by PCI Evol Biol. doi: https://doi.org/10.1101/2020.04.26.062265
[2] Reyserhove, L., Samaey, G., Muylaert, K., Coppé, V., Van Colen, W., and Decaestecker, E. (2017). A historical perspective of nutrient change impact on an infectious disease in Daphnia. Ecology, 98(11), 2784-2798. doi: https://doi.org/10.1002/ecy.1994
[3] Ebert, D., and Bull, J. J. (2003). Challenging the trade-off model for the evolution of virulence: is virulence management feasible?. Trends in microbiology, 11(1), 15-20. doi: https://doi.org/10.1016/S0966-842X(02)00003-3
[4] Houwenhuyse, S., Macke, E., Reyserhove, L., Bulteel, L., and Decaestecker, E. (2018). Back to the future in a petri dish: Origin and impact of resurrected microbes in natural populations. Evolutionary Applications, 11(1), 29-41. doi: https://doi.org/10.1111/eva.12538
[5] Day, T., and Gandon, S. (2007). Applying population‐genetic models in theoretical evolutionary epidemiology. Ecology Letters, 10(10), 876-888. doi: https://doi.org/10.1111/j.1461-0248.2007.01091.x
[6] Alizon, S., Hurford, A., Mideo, N., and Van Baalen, M. (2009). Virulence evolution and the trade‐off hypothesis: history, current state of affairs and the future. Journal of evolutionary biology, 22(2), 245-259. doi: https://doi.org/10.1111/j.1420-9101.2008.01658.x
[7] Alizon, S., de Roode, J. C., and Michalakis, Y. (2013). Multiple infections and the evolution of virulence. Ecology letters, 16(4), 556-567. doi: https://doi.org/10.1111/ele.12076
[8] Decaestecker, E., and King, K. (2019). Red queen dynamics. Reference module in earth systems and environmental sciences, 3, 185-192. doi: https://doi.org/10.1016/B978-0-12-409548-9.10550-0
[9] Kirk, D., Jones, N., Peacock, S., Phillips, J., Molnár, P. K., Krkošek, M., and Luijckx, P. (2018). Empirical evidence that metabolic theory describes the temperature dependency of within-host parasite dynamics. PLoS biology, 16(2), e2004608. doi: https://doi.org/10.1371/journal.pbio.2004608
[10] Frankel-Bricker, J., Song, M. J., Benner, M. J., and Schaack, S. (2019). Variation in the microbiota associated with Daphnia magna across genotypes, populations, and temperature. Microbial ecology, 1-12. doi: https://doi.org/10.1007/s00248-019-01412-9
| Experimental evolution of virulence and associated traits in a Drosophila melanogaster – Wolbachia symbiosis | David Monnin, Natacha Kremer, Caroline Michaud, Manon Villa, Hélène Henri, Emmanuel Desouhant, Fabrice Vavre | <p>Evolutionary theory predicts that vertically transmitted symbionts are selected for low virulence, as their fitness is directly correlated to that of their host. In contrast with this prediction, the Wolbachia strain wMelPop drastically reduces... |  | Evolutionary Ecology, Experimental Evolution, Species interactions | Ellen Decaestecker | | 2020-04-29 19:16:56 | View |
Multi-gene and lineage comparative assessment of the strength of selection in Hymenoptera
Recommended by Bertanne Visser based on reviews by Michael Lattorff and 1 anonymous reviewer
Genetic variation is the raw material for selection to act upon and the amount of genetic variation present within a population is a pivotal determinant of a population’s evolutionary potential. A large effective population size, i.e., the ideal number of individuals experiencing the same amount of genetic drift and inbreeding as an actual population, Ne (Wright 1931, Crow 1954), thus increases the probability of long-term survival of a population. However, natural populations, as opposed to theoretical ones, rarely adhere to the requirements of an ideal panmictic population (Sjödin et al. 2005). A range of circumstances can reduce Ne, including the structuring of populations (through space and time, as well as age and developmental stages) and inbreeding (Charlesworth 2009). In mammals, species with a larger body mass (as a proxy for lower Ne) were found to have a higher rate of nonsynonymous nucleotide substitutions (that alter the amino acid sequence of a protein), as well as radical amino acid substitutions (altering the physicochemical properties of a protein) (Popadin et al. 2007). In general, low effective population sizes increase the chance of mutation accumulation and drift, while reducing the strength of selection (Sjödin et al. 2005).
In this paper, Weyna and Romiguier (2021) set out to test if parasitism, body size, geographic range, and/or eusociality affect the strength of selection in Hymenoptera. Hymenoptera include the bees, wasps and ants and is an extraordinarily diverse order within the insects. It was recently estimated that Hymenoptera is the most speciose order of the animal kingdom (Forbes et al. 2018). Hymenoptera are further characterized by an impressive radiation of parasitic species, mainly parasitoids, that feed in or on a single host individual to complete their own development (Godfray 1994). All hymenopterans share the same sex determination system: haplo-diploidy, where unfertilized eggs are haploid males and fertilized eggs are diploid females. Compared to other animals, Hymenoptera further contain an impressive number of clades that evolved eusociality (Rehan and Toth 2015), in which societies show a clear division of labor for reproduction (i.e., castes) and cooperative brood care. Hymenopterans thus represent a diverse and interesting group of insects to investigate potential factors affecting strength of selection and Ne.
Using a previously published phylogenomic dataset containing 3256 genes and 169 hymenopteran species (Peters et al. 2017), Weyna and Romiguier (2021) estimated mean genomic dN/dS ratios (nonsynonymous to synonymous substitution rates) for each species and compared these values between parasitic and non-parasitic species, eusocial and solitary species, and in relation to body size, parasitoid-specific traits and geographic range, thought to affect the effective population size and strength of selection. The use of a large number of species, as well as several distinct traits is a clear asset of this study. The authors found no effect of body size, geographic range or parasitism (including a range of parasitoid-specific traits). There was an effect, however, of eusociality where dN/dS increased in three out of four eusocial lineages. Future studies including more independent evolutionary transitions to eusociality can lend further support that eusocial species indeed reduces the efficiency of selection. The most intriguing result was that for solitary and social bees, with high dN/dS ratios and a strong signature of relaxed selection (i.e., the elimination or reduction of a source of selection (Lahti et al. 2009). The authors suggest that the pollen-collecting behaviors of these species can constrain Ne, as pollen availability varies at both a spatial and temporal scale, requiring a large investment in foraging that may in turn limit reproductive output. It would be interesting to see if other pollen feeders, such as certain beetles, flies, butterflies and moths, as well as mites and spiders, experience relaxed selection as a consequence of the trade-off between energy investment in pollen foraging versus fecundity.
References
Charlesworth, B. (2009). Effective population size and patterns of molecular evolution and variation. Nature Reviews Genetics, 10(3), 195-205. doi: https://doi.org/10.1038/nrg2526
Crow, J. F. (1954) Statistics and Mathematics in Biology (eds Kempthorne, O., Bancroft, T. A., Gowen, J. W. & Lush, J. L.) 543–556 (Iowa State Univ. Press, Ames, Iowa)
Forbes, A. A., Bagley, R. K., Beer, M. A., Hippee, A. C., and Widmayer, H. A. (2018). Quantifying the unquantifiable: why Hymenoptera, not Coleoptera, is the most speciose animal order. BMC ecology, 18(1), 1-11. doi: https://doi.org/10.1186/s12898-018-0176-x
Godfray, H. C. J. (1994) Parasitoids: Behavioral and Evolutionary Ecology. Vol. 67, Princeton University Press, 1994. doi: https://doi.org/10.2307/j.ctvs32rmp
Lahti et al. (2009). Relaxed selection in the wild. Trends in ecology & evolution, 24(9), 487-496. doi: https://doi.org/10.1016/j.tree.2009.03.010
Peters et al. (2017). Evolutionary history of the Hymenoptera. Current Biology, 27(7), 1013-1018. doi: https://doi.org/10.1016/j.cub.2017.01.027
Popadin, K., Polishchuk, L. V., Mamirova, L., Knorre, D., and Gunbin, K. (2007). Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proceedings of the National Academy of Sciences, 104(33), 13390-13395. doi: https://doi.org/10.1073/pnas.0701256104
Rehan, S. M., and Toth, A. L. (2015). Climbing the social ladder: the molecular evolution of sociality. Trends in ecology & evolution, 30(7), 426-433. doi: https://doi.org/10.1016/j.tree.2015.05.004
Sjödin, P., Kaj, I., Krone, S., Lascoux, M., and Nordborg, M. (2005). On the meaning and existence of an effective population size. Genetics, 169(2), 1061-1070. doi: https://doi.org/10.1534/genetics.104.026799
Weyna, A., and Romiguier, J. (2021) Relaxation of purifying selection suggests low effective population size in eusocial Hymenoptera and solitary pollinating bees. bioRxiv, 2020.04.14.038893, ver. 5 peer-reviewed and recommended by PCI Evol Biol. doi: https://doi.org/10.1101/2020.04.14.038893
Wright, S. (1931). Evolution in Mendelian populations. Genetics, 16(2), 97-159.
| Relaxation of purifying selection suggests low effective population size in eusocial Hymenoptera and solitary pollinating bees | Arthur Weyna, Jonathan Romiguier | <p>With one of the highest number of parasitic, eusocial and pollinator species among all insect orders, Hymenoptera features a great diversity of lifestyles. At the population genetic level, such life-history strategies are expected to decrease e... | 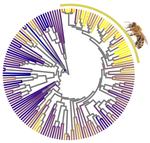 | Behavior & Social Evolution, Genome Evolution, Life History, Molecular Evolution, Population Genetics / Genomics | Bertanne Visser | | 2020-04-21 17:30:57 | View |
Sexual selection goes dynamic
Recommended by Michael D Greenfield based on reviews by Frédéric Guillaume and 1 anonymous reviewer
150 years after Darwin published ‘Descent of man and selection in relation to sex’ (Darwin, 1871), the evolutionary mechanism that he laid out in his treatise continues to fascinate us. Sexual selection is responsible for some of the most spectacular traits among animals, and plants, and it appeals to our interest in all things reproductive and sexual (Bell, 1982). In addition, sexual selection poses some of the more intractable problems in evolutionary biology: Its realm encompasses traits that are subject to markedly different selection pressures, particularly when distinct, yet associated, traits tend to be associated with males, e.g. courtship signals, and with females, e.g. preferences (cf. Ah-King & Ahnesjo, 2013). While separate, such traits cannot evolve independently of each other (Arnqvist & Rowe, 2005), and complex feedback loops and correlations between them are predicted (Greenfield et al., 2014). Traditionally, sexual selection has been modelled under simplifying assumptions, and quantitative genetic approaches that avoided evolutionary dynamics have prevailed. New computing methods may be able to free the field from these constraints, and a trio of theoreticians (Chevalier, De Coligny & Labonne 2020) describe here a novel application of a ‘demo-genetic agent (or individual) based model’, a mouthful hereafter termed DG-ABM, for arriving at a holistic picture of the sexual selection trajectory. The application is built on the premise that traits, e.g. courtship, preference, gamete investment, competitiveness for mates, can influence the genetic architecture, e.g. correlations, of those traits. In turn, the genetic architecture can influence the expression and evolvability of the traits. Much of this influence occurs via demographic features, i.e. social environment, generated by behavioral interactions during sexual advertisement, courtship, mate guarding, parental care, post-mating dispersal, etc.
The authors provide a lengthy verbal description of their model, specifying the genomic and behavioral parameters that can be set and how a ‘run’ may be initialized. There is a link to an internet site where users can then enter their own parameter values and begin exploring hypotheses. Back in the article several simulations illustrate simple tests; e.g. how gamete investment and preference jointly evolve given certain survival costs. One obvious test would have been the preference – courtship genetic correlation that represents the core of Fisherian runaway selection, and it is regrettable that it was not examined under a range of demographic parameters. As presented the author’s DG-ABM appears particularly geared toward mating systems in ‘higher’ vertebrates, where couples form during a discrete mating season and are responsible for most reproduction. It is not clear how applicable the model could be to a full range of mating systems and nuances, including those in arthropods and other invertebrates as well as plants.
What is the likely value of the DG-ABM for sexual selection researchers? We will not be able to evaluate its potential impact until readers with specialized understanding of a question and taxon begin exploring and comparing their results with prior expectations. Of course, lack of congruence with earlier predictions would not invalidate the model. Hopefully, some of these specialists will have opportunities for comparing results with pertinent empirical data.
References
Ah-King, M. and Ahnesjo, I. 2013. The ‘sex role’ concept: An overview and evaluation Evolutionary Biology, 40, 461-470. doi: https://doi.org/10.1007/s11692-013-9226-7
Arnqvist, G. and Rowe, L. 2005. Sexual Conflict. Princeton University Press, Princeton. doi: https://doi.org/10.1515/9781400850600
Bell, G. 1982. The Masterpiece of Nature: The Evolution and Genetics of Sexuality. University of California Press, Berkeley.
Chevalier, L., De Coligny, F. and Labonne, J. (2020) A demogenetic individual based model for the evolution of traits and genome architecture under sexual selection. bioRxiv, 2020.04.01.014514, ver. 4 peer-reviewed and recommended by PCI Evol Biol. doi: https://doi.org/10.1101/2020.04.01.014514
Darwin, C. 1871. The Descent of Man and Selection in Relation to Sex. J. Murray, London.
Greenfield, M.D., Alem, S., Limousin, D. and Bailey, N.W. 2014. The dilemma of Fisherian sexual selection: Mate choice for indirect benefits despite rarity and overall weakness of trait-preference genetic correlation. Evolution, 68, 3524-3536. doi: https://doi.org/10.1111/evo.12542
| A demogenetic agent based model for the evolution of traits and genome architecture under sexual selection | Louise Chevalier, François de Coligny, Jacques Labonne | <p>Sexual selection has long been known to favor the evolution of mating behaviors such as mate preference and competitiveness, and to affect their genetic architecture, for instance by favoring genetic correlation between some traits. Reciprocall... | 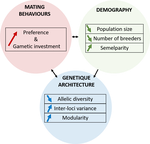 | Adaptation, Behavior & Social Evolution, Evolutionary Dynamics, Evolutionary Theory, Life History, Population Genetics / Genomics, Sexual Selection | Michael D Greenfield | | 2020-04-02 14:44:25 | View |
More intense symptoms, more treatment, more drug-resistance: coevolution of virulence and drug-resistance
Recommended by Ludek Berec based on reviews by 3 anonymous reviewers
Mathematical models play an essential role in current evolutionary biology, and evolutionary epidemiology is not an exception [1]. While the issues of virulence evolution and drug-resistance evolution resonate in the literature for quite some time [2, 3], the study by Alizon [4] is one of a few that consider co-evolution of both these traits [5]. The idea behind this study is the following: treating individuals with more severe symptoms at a higher rate (which appears to be quite natural) leads to an appearance of virulent drug-resistant strains, via treatment failure. The author then shows that virulence in drug-resistant strains may face different selective pressures than in drug-sensitive strains and hence proceed at different rates. Hence, treatment itself modulates evolution of virulence. As one of the reviewers emphasizes, the present manuscript offers a mathematical view on why the resistant and more virulent strains can be selected in epidemics. Also, we both find important that the author highlights that the topic and results of this study can be attributed to public health policies and development of optimal treatment protocols [6].
Mathematical models are simplified representations of reality, created with a particular purpose. It can be simple as well as complex, but even simple models can produce relatively complex and knitted results. The art of modelling thus lies not only in developing a model, but also in interpreting and unknitting the results. And this is what Alizon [4] indeed does carefully and exhaustively. Using two contrasting theoretical approaches to study co-evolution, the Price equation approach to study short-term evolution and the adaptive dynamics approach to study long-term evolution, Alizon [4] shows that a positive correlation between the rate of treatment and infection severity causes virulence in drug-sensitive strains to decrease. Clearly, no single model can describe and explain an examined system in its entirety, and even this aspect of the work is taken seriously. Many possible extensions of the study are laid out, providing a wide opportunity to pursue this topic even further. Personally, I have had an opportunity to read many Alizon’s papers and use, teach or discuss many of his models and results. All, including the current one, keep high standard and pursue the field of theoretical (evolutionary) epidemiology.
References
[1] Gandon S, Day T, Metcalf JE, Grenfell BT (2016) Forecasting epidemiological and evolutionary dynamics of infectious diseases. Trends Ecol Evol 31: 776-788. doi: https://doi.org/10.1016/j.tree.2016.07.010
[2] Berngruber TW, Froissart R, Choisy M, Gandon S (2013) Evolution of virulence in emerging epidemics. PLoS Pathog 9(3): e1003209. doi: https://doi.org/10.1371/journal.ppat.1003209
[3] Spicknall IH, Foxman B, Marrs CF, Eisenberg JNS (2013) A modeling framework for the evolution and spread of antibiotic resistance: literature review and model categorization. Am J Epidemiol 178: 508-520. doi: https://doi.org/10.1093/aje/kwt017
[4] Alizon S (2020) Treating symptomatic infections and the co-evolution of virulence and drug resistance. bioRxiv, 2020.02.29.970905, ver. 3 peer-reviewed and recommended by PCI Evol Biol. doi: https://doi.org/10.1101/2020.02.29.970905
[5] Carval D, Ferriere R (2010) A unified model for the coevolution of resistance, tolerance, and virulence. Evolution 64: 2988–3009. doi: https://doi.org/10.1111/j.1558-5646.2010.01035.x
[6] Read AF, T Day, and S Huijben (2011). The evolution of drug resistance and the curious orthodoxy of aggressive chemotherapy. Proc Natl Acad Sci USA 108 Suppl 2, 10871–7. doi: https://doi.org/10.1073/pnas.1100299108
| Treating symptomatic infections and the co-evolution of virulence and drug resistance | Samuel Alizon | <p>Antimicrobial therapeutic treatments are by definition applied after the onset of symptoms, which tend to correlate with infection severity. Using mathematical epidemiology models, I explore how this link affects the coevolutionary dynamics bet... | 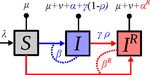 | Evolutionary Applications, Evolutionary Dynamics, Evolutionary Epidemiology, Evolutionary Theory | Ludek Berec | | 2020-03-04 10:18:39 | View |
Another step towards grasping the complexity of the environmental response of traits
Recommended by Benoit Pujol based on reviews by 2 anonymous reviewers
One can only hope that one day, we will be able to evaluate how the ecological complexity surrounding natural populations affects their ability to adapt. This is more like a long term quest than a simple scientific aim. Many steps are heading in the right direction. This paper by Monroe and colleagues (2021) is one of them.
Many ecological and genetic mechanisms shape the evolutionary potential of phenotypic trait variation and many of them involve environmental heterogeneity (Pujol et al 2018). To date, we cannot look into these ecological and genetic mechanisms without oversimplifying their effects. We often look into trait variation one trait at a time albeit the variation of multiple phenotypic traits is often linked at the genetic or environmental level. As a consequence, we put our conclusions at risk by not accounting for the reciprocal impacts of trait changes upon each other (Teplitsky et al 2014). We also usually restrict the study of a continuous gradient of environmental conditions to a few conditions because it would otherwise be impossible to model its environmental effect. As a consequence, we miss the full picture of the continuous often nonlinear phenotypic plastic response. Whether the trait undergo threshold effect changes thereby remains obscured to us. Collectively, these issues impede our ability to understand how selection shapes the ecological strategy of organisms under variable environments.
In this paper, Monroe and colleagues (2021) propose an original approach that raised to these two challenges. They analysed phenotypic plastic changes in response to a continuous environment in a multidimensional trait space, namely the response of Brachypodium plant developmental and physiological traits to a continuous gradient of soil moisture. They used dry down experimental treatments to produce the continuous soil moisture gradient and compared the plant capacity to use water between annual B. distachyon and perennial B. sylvaticum. Their results revealed the best mathematical functions that model the nonlinear curvature of the continuous plastic response of Brachypodium plants. This work reinforces our view that nonlinear plastic responses can result in greater or lesser trait values at any stage of the environmental gradient that were unexpected on the basis of linear predictors (Gienapp and Brommer 2014). Their findings also imply that different threshold responses characterize different genotypes. These could otherwise have been missed by a classical approach. By shedding light on unforeseen interactions between traits that make their correlation vary along the nonlinear response, they were able to describe more accurately Brachypodium ecological strategies and the changes in evolutionary constraints along the soil moisture gradient.
Their empirical approach allows to test what environmental conditions maximises the opportunity for selection to shape trait variation. For example, it revealed unforeseen divergence in potentially adaptive mechanisms or life history strategies – and not just trait values – between annual and perennial species of Brachypodium. Behind every environmental variation of the constraints to the future evolutionary change of multiple traits, we can expect that the evolutionary history of the populations shaped their trait genetic correlations. Investigating the nonlinear signature of adaptive evolution across continuous environments will get us into uncharted territory.
Our ability to predict the adaptive potential of species is limited. With their approach of continuous environmental gradients beyond linearity, Monroe and collaborators (2021) improve our understanding of plant phenotypic responses and open a brand new range of exciting developments. As they mention: "the opportunity for scaling up" their approach is big. To illustrate this prospect, I can easily think of an example: the quantitative genetic random regression model. This model allows to use any degree of genetic relatedness in a wild population to estimate the genetic variation of phenotypic plastic reaction norms (Nussey et al 2007, Pujol and Galaud 2013). However, in this approach, only a few modalities of the environmental gradient are used to model nonlinear phenotypic plastic responses. From there, it is rather intuitive. Combining the best of these two approaches (continuity of genetic relatedness in the wild & continuity of environmental gradient in experiments) could open ground breaking new perspectives in research.
References
Gienapp P. & J.E. Brommer. 2014. Evolutionary dynamics in response to climate change. In: Charmentier A, Garant D, Kruuk LEB, editors. Quantitative genetics in the wild. Oxford: Oxford University Press, Oxford. pp. 254–273. doi: https://doi.org/10.1093/acprof:oso/9780199674237.003.0015
Monroe, J. G., Cai, H., and Des Marais, D. L. (2020). Trait plasticity and covariance along a continuous soil moisture gradient. bioRxiv, 2020.02.17.952853, ver. 5 peer-reviewed and recommended by PCI Evol Biol. doi: https://doi.org/10.1101/2020.02.17.952853
Pujol et al. (2018). The missing response to selection in the wild. Trends in ecology & evolution, 33(5), 337-346. doi: https://doi.org/10.1016/j.tree.2018.02.007
Pujol, B., and Galaud, J. P. (2013). A practical guide to quantifying the effect of genes underlying adaptation in a mixed genomics and evolutionary ecology approach. Botany Letters, 160(3-4), 197-204. doi: https://doi.org/10.1080/12538078.2013.799045
Nussey, D. H., Wilson, A. J., and Brommer, J. E. (2007). The evolutionary ecology of individual phenotypic plasticity in wild populations. Journal of evolutionary biology, 20(3), 831-844. doi: https://doi.org/10.1111/j.1420-9101.2007.01300.x
Teplitsky et al. (2014). Assessing multivariate constraints to evolution across ten long-term avian studies. PLoS One, 9(3), e90444. doi: https://doi.org/10.1371/journal.pone.0090444
| Trait plasticity and covariance along a continuous soil moisture gradient | J Grey Monroe, Haoran Cai, David L Des Marais | <p>Water availability is perhaps the greatest environmental determinant of plant yield and fitness. However, our understanding of plant-water relations is limited because it is primarily informed by experiments considering soil moisture variabilit... |  | Phenotypic Plasticity | Benoit Pujol | | 2020-02-20 16:34:40 | View |
Further questions on the meaning of effective population size
Recommended by Martin Lascoux based on reviews by 3 anonymous reviewers
In spite of its name, the effective population size, Ne, has a complex and often distant relationship to census population size, as we usually understand it. In truth, it is primarily an abstract concept aimed at measuring the amount of genetic drift occurring in a population at any given time. The standard way to model random genetic drift in population genetics is the Wright-Fisher model and, with a few exceptions, definitions of the effective population size stems from it: “a certain model has effective population size, Ne, if some characteristic of the model has the same value as the corresponding characteristic for the simple Wright-Fisher model whose actual size is Ne” (Ewens 2004). Since Sewall Wright introduced the concept of effective population size in 1931 (Wright 1931), it has flourished and there are today numerous definitions of it depending on the process being examined (genetic diversity, loss of alleles, efficacy of selection) and the characteristic of the model that is considered. These different definitions of the effective population size were generally introduced to address specific aspects of the evolutionary process. One aspect that has been hotly debated since the first estimates of genetic diversity in natural populations were published is the so-called Lewontin’s paradox (1974). Lewontin noted that the observed variation in heterozygosity across species was much smaller than one would expect from the neutral expectations calculated with the actual size of the species.
In essence, what Galtier and Rousselle propose in their clever paper is to introduce a new approach to compare effective population sizes across species and thereby a new way to address Lewontin’s paradox. Classically, the effective population size in this type of comparative genomic studies is simply estimated from nucleotide diversity at putatively neutral sites using the equation relating levels of diversity (θ) to mutation rate per generation (μ) and effective population size, Ne, θ = 4Neμ. As Galtier and Rousselle point out there are many issues with this approach. In particular, although we can now estimate θ very precisely, we generally do not have a reliable estimate of the mutation rate, and the method rests on many, unwarranted, assumptions; for example that the population is at mutation-drift equilibrium. Instead they propose to estimate the effective population size from the load of segregating deleterious mutations which can be summarized by the ratio of nonsynonymous to synonymous mutations, πN/πS: small-Ne species are expected to accumulate more deleterious mutations and carry a higher load than large-Ne ones at selection/drift equilibrium (Ohta et al. 1973; Welch et al. 2008). At first glance, this suggestion seems counterintuitive since considering sites under selection undoubtedly adds a new layer of complexity to an already intricate situation. Indeed, one is now bringing to the brew another elusive object, namely, the Distribution of Fitness Effect of mutations (DFE). However, estimating Ne from the load of segregating deleterious mutations may actually simplify the situation in two important ways. First, using πN/πS does not require assumption about μ (as the mutation rate will cancel out in the ratio). Second, the ratio of nonsynonymous to synonymous nucleotide diversity, reaches equilibrium faster than the nucleotide diversity at synonymous site after a change in population size, so we can hope for less sensitivity to (often unknown) recent demographic history (Brandvain and Wright 2016).
Extending recent developments in the estimation of the DFE, Galtier and Rousselle eventually obtain estimates of the average deleterious effect, 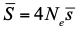 , where Ne is the effective population size and , where Ne is the effective population size and  is the mean fitness effect of non-synonymous mutations. Assuming further that distinct species share a common DFE and therefore a common is the mean fitness effect of non-synonymous mutations. Assuming further that distinct species share a common DFE and therefore a common  , they obtain estimates of the between species ratio of Ne from the between species ratio of , they obtain estimates of the between species ratio of Ne from the between species ratio of  . Applying their newly developed approach to various datasets they conclude that the power of drift varies by a factor of at least 500 between large-Ne (Drosophila) and small-Ne species (H. sapiens). This is an order of magnitude larger than what would be obtained by comparing estimates of the variation in neutral diversity. Hence the proposed approach seems to have gone some way in making Lewontin’s paradox less paradoxical. But, perhaps more importantly, as the authors tersely point out at the end of the abstract their results further questions the meaning of Ne parameters in population genetics. And arguably this could well be the most important contribution of their study and something that is badly needed. . Applying their newly developed approach to various datasets they conclude that the power of drift varies by a factor of at least 500 between large-Ne (Drosophila) and small-Ne species (H. sapiens). This is an order of magnitude larger than what would be obtained by comparing estimates of the variation in neutral diversity. Hence the proposed approach seems to have gone some way in making Lewontin’s paradox less paradoxical. But, perhaps more importantly, as the authors tersely point out at the end of the abstract their results further questions the meaning of Ne parameters in population genetics. And arguably this could well be the most important contribution of their study and something that is badly needed.
References
Brandvain Y, Wright SI (2016) The Limits of Natural Selection in a Nonequilibrium World. Trends in Genetics, 32, 201–210. doi: 10.1016/j.tig.2016.01.004
Ewens WJ (2010) Mathematical Population Genetics: Theoretical Introduction. Springer-Verlag New York Inc., New York, NY. doi: 10.1007/978-0-387-21822-9
Galtier N, Rousselle M (2020) How much does Ne vary among species? bioRxiv, 861849, ver. 4 peer-reviewed and recommended by PCI Evolutionary Biology. doi: 10.1101/861849
Lewontin RC (1974) The genetic basis of evolutionary change. Columbia University Press, New York.
Ohta T (1973) Slightly Deleterious Mutant Substitutions in Evolution. Nature, 246, 96–98. doi: 10.1038/246096a0
Welch JJ, Eyre-Walker A, Waxman D (2008) Divergence and Polymorphism Under the Nearly Neutral Theory of Molecular Evolution. Journal of Molecular Evolution, 67, 418–426. doi: 10.1007/s00239-008-9146-9
Wright S (1931) Evolution in Mendelian Populations. Genetics, 16, 97–159. url: https://www.genetics.org/content/16/2/97
| How much does Ne vary among species? | Nicolas Galtier, Marjolaine Rousselle | <p>Genetic drift is an important evolutionary force of strength inversely proportional to *Ne*, the effective population size. The impact of drift on genome diversity and evolution is known to vary among species, but quantifying this effect is a d... | 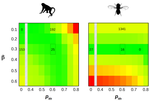 | Bioinformatics & Computational Biology, Genome Evolution, Molecular Evolution, Population Genetics / Genomics | Martin Lascoux | | 2019-12-08 00:11:00 | View |
How do invasion syndromes evolve? An experimental evolution approach using the ladybird Harmonia axyridis
Julien Foucaud, Ruth A. Hufbauer, Virginie Ravigné, Laure Olazcuaga, Anne Loiseau, Aurelien Ausset, Su Wang, Lian-Sheng Zang, Nicolas Lemenager, Ashraf Tayeh, Arthur Weyna, Pauline Gneux, Elise Bonnet, Vincent Dreuilhe, Bastien Poutout, Arnaud Estoup, Benoit Facon
https://doi.org/10.1101/849968
Selection on a single trait does not recapitulate the evolution of life-history traits seen during an invasion
Recommended by Inês Fragata and Ben Phillips based on reviews by 2 anonymous reviewers
Biological invasions are natural experiments, and often show that evolution can affect dynamics in important ways [1-3]. While we often think of invasions as a conservation problem stemming from anthropogenic introductions [4,5], biological invasions are much more commonplace than this, including phenomena as diverse as natural range shifts, the spread of novel pathogens, and the growth of tumors. A major question across all these settings is which set of traits determine the ability of a population to invade new space [6,7]. Traits such as: increased growth or reproductive rate, dispersal ability and ability to defend from predation often show large evolutionary shifts across invasion history [1,6,8]. Are such multi-trait shifts driven by selection on multiple traits, or a correlated response by multiple traits to selection on one? Resolving this question is important for both theoretical and practical reasons [9,10]. But despite the importance of this issue, it is not easy to perform the necessary manipulative experiments [9].
Foucaud et al. [11] tackled this issue by performing experimental evolution on source populations of the invasive ladybug Harmonia axyridis. The authors tested if selection on a single trait could generate correlated responses in other life history traits. Specifically, they used experimental evolution to impose divergent selection on female mass, and reproductive timing. After ten generations, they found that selection for weight did not affect almost any other life history trait. However, nine generations of selection for faster reproduction led to correlated phenotypic changes in developmental, reproduction and survival rate of populations, although not always in the direction we might have expected. Despite this correlated response, none of their selected lines were able to fully recapitulate the trait shifts seen in natural invasions of this species. This implies that selection during natural invasions is operating on multiple traits; a finding in agreement with our growing understanding of how selection acts during introduction and invasion [12,13].
Populations undergoing a colonization process may also be subject to a multitude of different selective pressures [14,15]. The authors expanded their work in this direction by testing whether food availability alters the observed correlations between life history traits. The pervasiveness of genotype by environment interactions observed also points to a role for multiple selective pressures in shaping the suite of life-history shifts observed in wild ladybug populations. The work from Foucaud and colleagues [11] adds to a small but growing list of important studies that use experimental evolution to investigate how life-history traits evolve, and how they evolve during invasions in particular.
References
[1] Sakai, A.K., Allendorf, F.W., Holt, J.S. et al. (2001). The population biology of invasive species. Annual review of ecology and systematics, 32(1), 305-332. doi: 10.1146/annurev.ecolsys.32.081501.114037
[2] Hairston Jr, N. G., Ellner, S. P., Geber, M. A., Yoshida, T. and Fox, J. A. (2005). Rapid evolution and the convergence of ecological and evolutionary time. Ecology letters, 8(10), 1114-1127. doi: 10.1111/j.1461-0248.2005.00812.x
[3] Chuang, A. and Peterson, C. R. (2016). Expanding population edges: theories, traits, and trade‐offs. Global change biology, 22(2), 494-512. doi: 10.1111/gcb.13107
[4] Whitney, K. D. and Gabler, C. A. (2008). Rapid evolution in introduced species,‘invasive traits’ and recipient communities: challenges for predicting invasive potential. Diversity and Distributions, 14(4), 569-580. doi: 10.1111/j.1472-4642.2008.00473.x
[5] Catullo, R. A., Llewelyn, J., Phillips, B. L. and Moritz, C. C. (2019). The Potential for Rapid Evolution under Anthropogenic Climate Change. Current Biology, 29(19), R996-R1007. doi: 10.1016/j.cub.2019.08.028
[6] Suarez, A. V. and Tsutsui, N. D. (2008). The evolutionary consequences of biological invasions. Molecular Ecology, 17(1), 351-360. doi: 10.1111/j.1365-294X.2007.03456.x
[7] Deforet, M., Carmona-Fontaine, C., Korolev, K. S. and Xavier, J. B. (2019). Evolution at the edge of expanding populations. The American Naturalist, 194(3), 291-305. doi: 10.1086/704594
[8] Phillips, B. L., Brown, G. P., and Shine, R. (2010). Life‐history evolution in range‐shifting populations. Ecology, 91(6), 1617-1627. doi: 10.1890/09-0910.1
[9] Colautti, R. I. and Lau, J. A. (2015). Contemporary evolution during invasion: evidence for differentiation, natural selection, and local adaptation. Molecular ecology, 24(9), 1999-2017. doi: 10.1111/mec.13162
[10] Szűcs, M., Melbourne, B. A., Tuff, T., Weiss‐Lehman, C. and Hufbauer, R. A. (2017). Genetic and demographic founder effects have long‐term fitness consequences for colonising populations. Ecology Letters, 20(4), 436-444. doi: 10.1111/ele.12743
[11] Foucaud, J., Hufbauer, R. A., Ravigné, V., Olazcuaga, L., Loiseau, A., Ausset, A., Wang, S., Zang, L.-S., Lemenager, N., Tayeh, A., Weyna, A., Gneux, P., Bonnet, E., Dreuilhe, V., Poutout, B., Estoup, A. and Facon, B. (2020). How do invasion syndromes evolve? An experimental evolution approach using the ladybird Harmonia axyridis. bioRxiv, 849968 ver. 4 peer-reviewed and recommended by PCI Evolutionary Biology. doi: 10.1101/849968
[12] Simons, A. M. (2003). Invasive aliens and sampling bias. Ecology Letters, 6(4), 278-280. doi: 10.1046/j.1461-0248.2003.00430.x
[13] Phillips, B. L. and Perkins, T. A. (2019). Spatial sorting as the spatial analogue of natural selection. Theoretical Ecology, 12(2), 155-163. doi: 10.1007/s12080-019-0412-9
[14] Lavergne, S. and Molofsky, J. (2007). Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proceedings of the National Academy of Sciences, 104(10), 3883-3888. doi: 10.1073/pnas.0607324104
[15] Moran, E. V. and Alexander, J. M. (2014). Evolutionary responses to global change: lessons from invasive species. Ecology Letters, 17(5), 637-649. doi: 10.1111/ele.12262
| How do invasion syndromes evolve? An experimental evolution approach using the ladybird Harmonia axyridis | Julien Foucaud, Ruth A. Hufbauer, Virginie Ravigné, Laure Olazcuaga, Anne Loiseau, Aurelien Ausset, Su Wang, Lian-Sheng Zang, Nicolas Lemenager, Ashraf Tayeh, Arthur Weyna, Pauline Gneux, Elise Bonnet, Vincent Dreuilhe, Bastien Poutout, Arnaud Est... | <p>Experiments comparing native to introduced populations or distinct introduced populations to each other show that phenotypic evolution is common and often involves a suit of interacting phenotypic traits. We define such sets of traits that evol... |  | Adaptation, Evolutionary Applications, Experimental Evolution, Life History, Quantitative Genetics | Inês Fragata | | 2019-11-29 07:07:00 | View |
Phylogenomic approaches reveal how a climatic inversion and glacial refugia shape patterns of diversity in an African rain forest tree species
Andrew J. Helmstetter, Biowa E. N. Amoussou, Kevin Bethune, Narcisse G. Kandem, Romain Glèlè Kakaï, Bonaventure Sonké, Thomas L. P. Couvreur
https://doi.org/10.1101/807727
Remarkable insights into processes shaping African tropical tree diversity
Recommended by Michael Pirie based on reviews by Miguel de Navascués, Lars Chatrou and Oscar Vargas based on reviews by Miguel de Navascués, Lars Chatrou and Oscar Vargas
Tropical biodiversity is immense, under enormous threat, and yet still poorly understood. Global climatic breakdown and habitat destruction are impacting on and removing this diversity before we can understand how the biota responds to such changes, or even fully appreciate what we are losing [1]. This is particularly the case for woody shrubs and trees [2] and for the flora of tropical Africa [3].
Helmstetter et al. [4] have taken a significant step to improve our understanding of African tropical tree diversity in the context of past climatic change. They have done so by means of a remarkably in-depth analysis of one species of the tropical plant family Annonaceae: Annickia affinis [5]. A. affinis shows a distribution pattern in Africa found in various plant (but interestingly not animal) groups: a discontinuity between north and south of the equator [6]. There is no obvious physical barrier to cause this discontinuity, but it does correspond with present day distinct northern and southern rainy seasons. Various explanations have been proposed for this discontinuity, set out as hypotheses to be tested in this paper: climatic fluctuations resulting in changes in plant distributions in the Pleistocene, or differences in flowering times or in ecological niche between northerly and southerly populations. These explanations are not mutually exclusive, but they can be tested using phylogenetic inference – if you can sample variable enough sequence data from enough individuals – complemented with analysis of ecological niches and traits.
Using targeted sequence capture, the authors amassed a dataset representing 351 nuclear markers for 112 individuals of A. affinis. This dataset is impressive for a number of reasons: First, sampling such a species across such a wide range in tropical Africa presents numerous challenges of itself. Second, the technical achievement of using this still relatively new sequencing technique with a custom set of baits designed specifically for this plant family [7] is also considerable. The result is a volume of data that just a few years ago would not have been feasible to collect, and which now offers the possibility to meaningfully analyse DNA sequence variation within a species across numerous independent loci of the nuclear genome. This is the future of our research field, and the authors have ably demonstrated some of its possibilities.
Using this data, they performed on the one hand different population genetic clustering approaches, and on the other, different phylogenetic inference methods. I would draw attention to their use and comparison of coalescence and network-based approaches, which can account for the differences between gene trees that might be expected between populations of a single species. The results revealed four clades and a consistent sequence of divergences between them. The authors inferred past shifts in geographic range (using a continuous state phylogeographic model), depicting a biogeographic scenario involving a dispersal north over the north/south discontinuity; and demographic history, inferring in some (but not all) lineages increases in effective population size around the time of the last glacial maximum, suggestive of expansion from refugia. Using georeferenced specimen data, they compared ecological niches between populations, discovering that overlap was indeed smallest comparing north to south. Just the phenology results were effectively inconclusive: far better data on flowering times is needed than can currently be harvested from digitised herbarium specimens.
Overall, the results add to the body of evidence for the impact of Pleistocene climatic changes on population structure, and for niche differences contributing to the present day north/south discontinuity. However, they also paint a complex picture of idiosyncratic lineage-specific responses, even within a single species. With the increasing accessibility of the techniques used here we can look forward to more such detailed analyses of independent clades necessary to test and to expand on these conclusions, better to understand the nature of our tropical plant diversity while there is still opportunity to preserve it for future generations.
References
[1] Mace, G. M., Gittleman, J. L., and Purvis, A. (2003). Preserving the Tree of Life. Science, 300(5626), 1707–1709. doi: 10.1126/science.1085510
[2] Humphreys, A. M., Govaerts, R., Ficinski, S. Z., Nic Lughadha, E., and Vorontsova, M. S. (2019). Global dataset shows geography and life form predict modern plant extinction and rediscovery. Nature Ecology and Evolution, 3(7), 1043–1047. doi: 10.1038/s41559-019-0906-2
[3] Stévart, T., Dauby, G., Lowry, P. P., Blach-Overgaard, A., Droissart, V., Harris, D. J., Mackinder, B. A., Schatz, G. E., Sonké, B., Sosef, M. S. M., Svenning, J. C., Wieringa, J. J., and Couvreur, T. L. P. (2019). A third of the tropical African flora is potentially threatened with extinction. Science Advances, 5(11), eaax9444. doi: 10.1126/sciadv.aax9444
[4] Helmstetter, A. J., Amoussou, B. E. N., Bethune, K., Kandem, N. G., Kakaï, R. G., Sonké, B., and Couvreur, T. L. P. (2020). Phylogenomic approaches reveal how a climatic inversion and glacial refugia shape patterns of diversity in an African rain forest tree species. BioRxiv, 807727, ver. 3 peer-reviewed and recommended by PCI Evolutionary Biology. doi: 10.1101/807727
[5] Versteegh, C. P. C., and Sosef, M. S. M. (2007). Revision of the African genus Annickia (Annonaceae). Systematics and Geography of Plants, 77, 91–118.
[6] Hardy, O. J., Born, C., Budde, K., Daïnou, K., Dauby, G., Duminil, J., Ewédjé, E.-E. B. K., Gomez, C., Heuertz, M., Koffi, G. K., Lowe, A. J., Micheneau, C., Ndiade-Bourobou, D., Piñeiro, R., and Poncet, V. (2013). Comparative phylogeography of African rain forest trees: A review of genetic signatures of vegetation history in the Guineo-Congolian region. Comptes Rendus Geoscience, 345(7), 284-296. doi: 10.1016/j.crte.2013.05.001
[7] Couvreur, T. L. P., Helmstetter, A. J., Koenen, E. J. M., Bethune, K., Brandão, R. D., Little, S. A., Sauquet, H., and Erkens, R. H. J. (2019). Phylogenomics of the Major Tropical Plant Family Annonaceae Using Targeted Enrichment of Nuclear Genes. Frontiers in Plant Science, 9. doi: 10.3389/fpls.2018.01941
| Phylogenomic approaches reveal how a climatic inversion and glacial refugia shape patterns of diversity in an African rain forest tree species | Andrew J. Helmstetter, Biowa E. N. Amoussou, Kevin Bethune, Narcisse G. Kandem, Romain Glèlè Kakaï, Bonaventure Sonké, Thomas L. P. Couvreur | <p>The world’s second largest expanse of tropical rain forest is in Central Africa and it harbours enormous species diversity. Population genetic studies have consistently revealed significant structure across central African rain forest plants, i... | 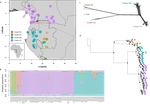 | Evolutionary Dynamics, Phylogeography & Biogeography | Michael Pirie | | 2019-10-29 15:19:36 | View |
Evolution and genetic architecture of disassortative mating at a locus under heterozygote advantage
Ludovic Maisonneuve, Mathieu Joron, Mathieu Chouteau and Violaine Llaurens
https://doi.org/10.1101/616409
Evolutionary insights into disassortative mating and its association to an ecologically relevant supergene
Recommended by Charles Mullon based on reviews by Tom Van Dooren and 2 anonymous reviewers based on reviews by Tom Van Dooren and 2 anonymous reviewers
Heliconius butterflies are famous for their colorful wing patterns acting as a warning of their chemical defenses [1]. Most species are involved in Müllerian mimicry assemblies, as predators learn to associate common wing patterns with unpalatability and preferentially target rare variants. Such positive-frequency dependent selection homogenizes wing patterns at different localities, and in several species, all individuals within a community belong to the same morph [2]. In this respect, H. numata stands out. This species shows stable local polymorphism across multiple localities, with local populations home to up to seven distinct morphs [2]. Although a balance between migration and local positive-frequency dependent selection can allow some degree of local polymorphism, theory suggests that this occurs only when migration is within a narrow window [3].
One factor that potentially enhances local polymorphism in H. numata is disassortative mating. Mate choice assays have in fact revealed that females of this species tend to reject males with the same wing pattern [4]. However the evolution of such mating behavior and its effect on polymorphism remain unclear when selection is locally positive-frequency dependent. Using a mathematical model, Maisonneuve et al. [5] clarify the conditions that favor the evolution of disassortative mating in the complicated system of H. numata. In particular, they investigate whether the genetic basis of wing colour can favor the emergence of disassortative mating. Variation in wing pattern in H. numata is controlled by the supergene P, which is a single genomic region harboring multiple protein coding genes that have ceased to recombine due to chromosomal inversions [6]. If such remarkable genetic configuration allows for the co-adaptation of multiple loci participating to a complex phenotype such as wing color pattern, the absence of recombination can also result in the accumulation of deleterious mutations [7]. In fact, alleles at the P locus have been associated with a recessive genetic load, leading to a fitness advantage for heterozygotes at this locus [8]. Can this fitness advantage to heterozygotes lead to the evolution of disassortative mating? And if so, can such evolution lead to the maintenance of local polymorphism in spite of strong positive frequency-dependent selection?
To investigate these questions, Maisonneuve et al. [5] model evolution at two loci, one is the P locus for wing pattern, and the other influences mating behavior. The population is divided among two connected patches that differ in their butterfly communities, so that different alleles at the P locus are favored by positive frequency-dependent selection in different patches. The different alleles at the P locus are ordered in dominance relationships such that the most dominant over wing color pattern are also those with the highest load. By tracking the dynamics of haplotype frequencies in the population, the authors first show that disassortative mating readily evolves via the invasion of an allele causing females carrying it to reject males that resemble them phenotypically. Such “self-referencing” mechanism of mate choice, however, has never been reported and has been argued to be rare due to its complicated nature [9].
Maisonneuve et al. [5] then compare the evolution of disassortative mating via two alternative mechanisms: attraction and rejection. In these cases, alleles at the mating locus determine attraction to or rejection of specific phenotypes (e.g., under attraction rule, allele “B” encodes attraction to males with phenotype B). With the P and mating loci fully linked, disassortative mating can evolve under all three mechanisms (self-referencing, attraction and rejection), but tends to be less prevalent at equilibrium under attraction rule. This in turn results in the maintenance of less genetic variation under attraction compared to the other mating mechanisms. The loss of variation that occurs under attraction rules is due to a combination of dominance relationships between alleles at the P locus and the searching cost to females in finding rare types of males. When a particular wing pattern, say B, is only expressed in homozygotic form, B males are relatively rare. Females that carry the allele at the mating locus causing them to be attracted to such males then suffer a fitness cost due to lost mating opportunities. This mating allele is therefore purged, and in turn so is the recessive allele for B phenotype at the P locus. Under self-referencing and rejection rules, however, choosy females only reject males of a specific phenotype. They can therefore potentially mate with larger pool of males than females attracted to a single type. As a result, self-referencing and rejection rules are less sensitive to demographic effects and so are more conducive to disassortative mating evolution.
In their final analysis, Maisonneuve et al. [5] investigate the influence of recombination among the P and mating loci. They show that recombination has different effects on disassortative mating evolution depending on the mechanism of mate choice. Under the self-referencing rule, loose linkage leads to higher levels of disassortative mating and polymorphism than when linkage is tight. Under attraction or rejection rule, however, even very limited recombination completely inhibits the evolution of disassortative mating. This is because, with alleles at the mating locus coding for attraction/rejection to specific males, recombination breaks the association between the P and mating loci necessary for disassortative mating. By contrast, disassortative mating via a self-referencing rule does not depend on the linkage among the P and mating loci: females choose males that are different to themselves independently from the alleles they carry at the P locus.
Taken together, Maisonneuve et al.’s analyses [5] show that disassortative mating can readily evolve in a system like H. numata, but that this evolution depends on the genetic architecture of mating behavior. The architectures that are more conducive to the evolution of disassortative mating are: (1) epistatic interactions among the P and mating loci such that females are able to recognize their own phenotype and base their mating decision upon this information (self-referencing rule); and (2) full linkage among the P supergene and a mating locus that triggers rejection of a specific color pattern. While the mechanisms behind disassortative mating remain to be elucidated, assortative mating seems to rely on alleles triggering attraction to specific cues with variation in attraction and cues linked together [10]. These observations support the notion that disassortative mating is due to alleles causing rejection, in tight linkage to the P locus. If so, mating loci would in fact be part of the P supergene, thus controlling not only intricate wing color pattern but also mating behavior.
Beyond the specific system of H. numata, Maisonneuve et al.’s study [5] helps understand the evolution of disassortative mating and its association with the genetic architecture of correlated traits. In particular, Maisonneuve et al. [5] expands the role of supergenes for ecologically relevant traits to mating behavior, further bolstering the relevance of these remarkable genetic elements in the maintenance of variation in complex and elaborate phenotypes.
References
[1] Merrill, R M, K K Dasmahapatra, J W Davey, D D Dell'Aglio, J J Hanly, B Huber, C D Jiggins, et al. (2015). The Diversification of Heliconius butterflies: What Have We Learned in 150 Years? Journal of Evolutionary Biology 28 (8), 1417–38. https://doi.org/10.1111/jeb.12672.
[2] Joron M, IR Wynne, G Lamas, and J Mallet (1999). Variable selection and the coexistence of multiple mimetic forms of the butterfly Heliconius numata. Evolutionary Ecology 13, 721– 754. https://doi.org/10.1023/A:1010875213123
[3] Joron M and Y Iwasa (2005). The evolution of a Müllerian mimic in a spatially distributed community. Journal of Theoretical Biology 237, 87–103. https://doi.org/10.1016/j.jtbi.2005.04.005
[4] Chouteau M, V Llaurens, F Piron-Prunier, and M Joron (2017). Polymorphism at a mimicry su- pergene maintained by opposing frequency-dependent selection pressures. Proceedings of the National Academy of Sciences 114, 8325–8329. https://doi.org/10.1073/pnas.1702482114
[5] Maisonneuve, L, Chouteau, M, Joron, M and Llaurens, V. (2020). Evolution and genetic architecture of disassortative mating at a locus under heterozygote advantage. bioRxiv, 616409, ver. 9 peer-reviewed and recommended by PCI Evolutionary Biology. https://doi.org/10.1101/616409
[6] Joron M, L Frezal, RT Jones, NL Chamberlain, SF Lee, CR Haag, A Whibley, M Becuwe, SW Baxter, L Ferguson, et al. (2011). Chromosomal rearrangements maintain a polymorphic super- gene controlling butterfly mimicry. Nature 477, 203. https://doi.org/10.1038/nature10341
[7] Schwander T, R Libbrecht, and L Keller (2014). Supergenes and Complex Phenotypes.” Current Biology. 24 (7), 288–94. https://doi.org/10.1016/j.cub.2014.01.056.
[8] Jay P, M Chouteau, A Whibley, H Bastide, V Llaurens, H Parrinello, and M Joron (2019). Mutation accumulation in chromosomal inversions maintains wing pattern polymorphism in a butterfly. bioRxiv. https://doi.org/ 10.1101/736504.
[9] Kopp M, MR Servedio, TC Mendelson, RJ Safran, RL Rodrıguez, ME Hauber, EC Scordato, LB Symes, CN Balakrishnan, DM Zonana, et al. (2018). Mechanisms of assortative mating in speciation with gene flow: connecting theory and empirical research. The American Naturalist 191, 1–20. https://doi.org/10.1086/694889
[10] Merrill RM, P Rastas, SH Martin, MC Melo, S Barker, J Davey, WO McMillan, and CD Jiggins (2019). Genetic dissection of assortative mating behavior. PLoS biology 17, e2005902. https://doi.org/10.1371/journal.pbio.2005902
| Evolution and genetic architecture of disassortative mating at a locus under heterozygote advantage | Ludovic Maisonneuve, Mathieu Joron, Mathieu Chouteau and Violaine Llaurens | <p>The evolution of mate preferences may depend on natural selection acting on the mating cues and on the underlying genetic architecture. While the evolution of assortative mating with respect to locally adapted traits has been well-characterized... |  | Evolutionary Theory, Population Genetics / Genomics, Reproduction and Sex, Sexual Selection | Charles Mullon | | 2019-10-29 09:55:18 | View |
Epistasis and the evolution of selfing
Recommended by Sylvain Gandon based on reviews by Nick Barton and 1 anonymous reviewer
The evolution of selfing results from a balance between multiple evolutionary forces. Selfing provides an "automatic advantage" due to the higher efficiency of selfers to transmit their genes via selfed and outcrossed offspring. Selfed offspring, however, may suffer from inbreeding depression. In principle the ultimate evolutionary outcome is easy to predict from the relative magnitude of these two evolutionary forces [1,2]. Yet, several studies explicitly taking into account the genetic architecture of inbreeding depression noted that these predictions are often too restrictive because selfing can evolve in a broader range of conditions [3,4].
The present work by Abu Awad and Roze [5] provides an analytic understanding of these results. Abu Awad and Roze analyse the evolution of selfing in a multilocus model where some loci are coding for selfing while others are under direct selection. The evolution of selfing depends on (i) the classical benefit of selfing (automatic advantage), (ii) the cost of selfing due to inbreeding depression, (iii) the association between the loci coding for selfing and the loci under direct selection (likely to be positive because selfing is expected to be found in better purged genetic backgrounds) and (iv) the association between the loci coding for selfing and the linkage between loci under selection (this final term depends on the magnitude and the type of epistasis). Because these last two terms depend on genetic associations they are expected to play in when selection is strong and recombination is small. These last two terms explain why selfing is evolving under a range of conditions which is broader than predicted by earlier theoretical models. The match between the approximations for the different terms acting on the evolution of selfing and individual based simulations (for different fitness landscapes) is very convincing. In particular, this analysis also yields new results on the effect of different types of epistasis on inbreeding depression.
Another remarkable and important feature of this work is its readability. The analysis of multilocus models rely on several steps and approximations that often result in overwhelmingly complex papers. Abu Awad and Roze’s paper [5] is dense but it provides a very clear and comprehensive presentation of the interplay between multiple evolutionary forces acting on the evolution of selfing.
References
[1] Holsinger, K. E., Feldman, M. W., and Christiansen, F. B. (1984). The evolution of self-fertilization in plants: a population genetic model. The American Naturalist, 124(3), 446-453. doi: 10.1086/284287
[2] Lande, R., and Schemske, D. W. (1985). The evolution of self‐fertilization and inbreeding depression in plants. I. Genetic models. Evolution, 39(1), 24-40. doi: 10.1111/j.1558-5646.1985.tb04077.x
[3] Charlesworth, D., Morgan, M. T., and Charlesworth, B. (1990). Inbreeding depression, genetic load, and the evolution of outcrossing rates in a multilocus system with no linkage. Evolution, 44(6), 1469-1489. doi: 10.1111/j.1558-5646.1990.tb03839.x
[4] Uyenoyama, M. K., and Waller, D. M. (1991). Coevolution of self-fertilization and inbreeding depression I. Mutation-selection balance at one and two loci. Theoretical population biology, 40(1), 14-46. doi: 10.1016/0040-5809(91)90045-H
[5] Abu Awad, D. and Roze, D. (2020). Epistasis, inbreeding depression and the evolution of self-fertilization. bioRxiv, 809814, ver. 4 peer-reviewed and recommended by PCI Evol Biol. doi: 10.1101/809814
| Epistasis, inbreeding depression and the evolution of self-fertilization | Diala Abu Awad and Denis Roze | <p>Inbreeding depression resulting from partially recessive deleterious alleles is thought to be the main genetic factor preventing self-fertilizing mutants from spreading in outcrossing hermaphroditic populations. However, deleterious alleles may... | 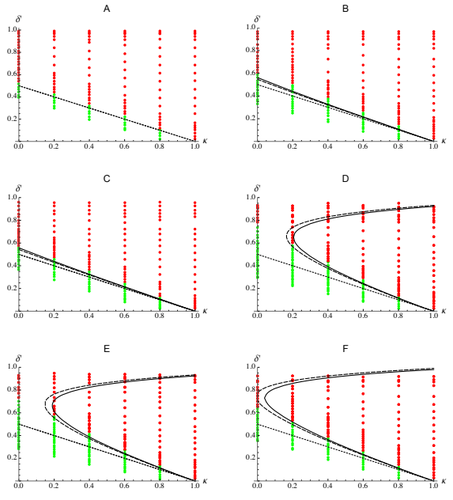 | Evolutionary Theory, Quantitative Genetics, Reproduction and Sex | Sylvain Gandon | | 2019-10-18 09:29:41 | View |

















 based on reviews by Miguel de Navascués, Lars Chatrou and Oscar Vargas
based on reviews by Miguel de Navascués, Lars Chatrou and Oscar Vargas


 based on reviews by Tom Van Dooren and 2 anonymous reviewers
based on reviews by Tom Van Dooren and 2 anonymous reviewers





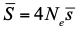 , where Ne is the effective population size and
, where Ne is the effective population size and  is the mean fitness effect of non-synonymous mutations. Assuming further that distinct species share a common DFE and therefore a common
is the mean fitness effect of non-synonymous mutations. Assuming further that distinct species share a common DFE and therefore a common  . Applying their newly developed approach to various datasets they conclude that the power of drift varies by a factor of at least 500 between large-Ne (Drosophila) and small-Ne species (H. sapiens). This is an order of magnitude larger than what would be obtained by comparing estimates of the variation in neutral diversity. Hence the proposed approach seems to have gone some way in making Lewontin’s paradox less paradoxical. But, perhaps more importantly, as the authors tersely point out at the end of the abstract their results further questions the meaning of Ne parameters in population genetics. And arguably this could well be the most important contribution of their study and something that is badly needed.
. Applying their newly developed approach to various datasets they conclude that the power of drift varies by a factor of at least 500 between large-Ne (Drosophila) and small-Ne species (H. sapiens). This is an order of magnitude larger than what would be obtained by comparing estimates of the variation in neutral diversity. Hence the proposed approach seems to have gone some way in making Lewontin’s paradox less paradoxical. But, perhaps more importantly, as the authors tersely point out at the end of the abstract their results further questions the meaning of Ne parameters in population genetics. And arguably this could well be the most important contribution of their study and something that is badly needed. 







